REVIEW
miRNA biogenesis and functioning: 30 years since their discovery
1 Chazov National Medical Research Center of Cardiology, Moscow, Russia
2 Pirogov Russian National Research Medical University
The role of miRNAs (small non-coding RNAs) in regulation of gene expression is reported. By binding with target mRNAs miRNAs control expression of the genes encoding these mRNAs at post-transcriptional level taking part in physiological and pathological processes, from embryogenesis to neoplastic disorders. Various research teams have been studying the miRNA functions and mechanisms of action since the discovery of these molecules in 1993. The paper reports miRNA biogenesis pathways, modes of interaction between miRNAs and target mRNAs, and the mechanisms underlying suppression of translation and mRNA degradation. The results of numerous studies have shown that miRNAs can be used in medicine as biomarkers for diagnostic and prognostic purposes. Developments in miRNA therapeutics hold promise for the treatment of diseases, in which gene dysregulation plays a key role.
Keywords: diagnostics, miRNA, therapy, regulation of gene expression, miRNA biogenesis, prognosis of disease progression
Funding: the study was conducted within the framework of the State Assignment of the Chazov National Medical Research Center of Cardiology (No. 124020200013-3).
Author contribution: Pisklova MV — literature data acquisition, analysis and systematization, planning and writing the manuscript draft, selecting drawings; Baulina NM, Favorova OO — planning, manuscript structuring, editing; Matveeva NA — literature data analysis and systematization, manuscript writing.
Correspondence should be addressed: Maria V. Pisklova
Akademika Chazova, 15А, Moscow, 121552, Russia; ur.liam@airam_avolksip
In 1993, research groups of American biologists Victor Ambros and Gary Ruvkun reported a short RNA of the nematode Caenorhabditis elegans, which did not encode any protein, but played a key role in regulation of the nematode development via suppression of the LIN-14 protein mRNA translation [1, 2]. This small RNA referred to as lin-4 became the first discovered short RNA having regulatory properties. In 2000, the second short RNA, let-7, having the same mechanism of action, was found in Caenorhabditis elegans by Gary Ruvkun and collegues [3]. From that moment began extensive study of the new type of small non-coding RNAs, 21–24 nucleotides in length, referred to as microRNAs (miRNAs), the discovery of which significantly influences the established ideas about regulation of gene functioning. While before this discovery the main mechanisms underlying regulation of transcription and RNA splicing implemented by specific proteins in the nucleus were known, currently this knowledge is complemented by the idea about further control of gene expression in the cytoplasm through miRNAs. This mechanism is of fundamental importance for the development and functioning of all cell types, it involves regulation of the target genes expression through the interaction with their mRNAs at post-transcriptional level. The presence of miRNA molecules was reported for various eukaryotic species, including plants and animals, as well as for some viruses [4]. In 2024, V. Ambros and G. Ruvkun were awarded the Nobel Prize in Physiology or Medicine “for the discovery of microRNA and its role in post-transcriptional gene regulation”.
In recent years, it was found that numerous miRNAs are important for regulation of almost all physiological and disease processes: from early stages of embryogenesis to body’s ageing and death. Let us consider the current state of knowledge about miRNA biogenesis and functioning.
miRNA abundance and nomenclature
According to the last version of miRBase, the miRNA sequence and annotation database, by 17.03.2024 a total of 48,860 mature miRNAs have been found in 271 species, of those 2654 have been identified in human body. The number of miRNAs detected continues to grow. The range of miRNAs present in this or that organism depends directly on its structural complexity [5]. The evolutionarily related miRNAs are grouped into various families (there are 267 families in humans), the members of which have highly homologous sequences and some common targets. Highly conservative nature of the nucleotide sequences of some miRNAs (primarily in the target mRNA binding region) in phylogenesis has been demonstrated. In general, miRNA evolution is closely linked to evolution of target genes [5]. miRNAs are numbered consecutively as they are discovered. The experimentally confirmed mature miRNAs are assigned a number, which is attached to the “miR” prefix through a hyphen (for example, miR-499). The "miR" can be preceded by a threeletter abbreviation indicating the species (for example, “hsa” for Homo sapiens, “mmu” for Mus musculus). miRNAs identical in the sequence but transcribed from different regions of the genome are added a numeric suffix with an ordinal number, for example, hsa-miR-219-1 and hsa-miR-219-2. A letter suffix is added to the names of miRNAs, the sequences of which differ slightly (by 1–2 nucleotides), for example, hsa-miR-130a and hsa-miR-130b; they form miRNA families. miRNAs, the genes of which are located physically close to each other and are often transcribed as a single unit, are grouped in clusters, which are named either based on the lowest miRNA number in the cluster (for example, cluster miR-17), or based on the lowest and highest miRNA numbers written through a hyphen (cluster miR-17-92 consisting of miR-17, miR-91, miR-18, miR-19, miR-19b, miR-20, and miR-92) [6].
Genes and miRNA biogenesis
Depending on their localization relative to the genome components, miRNA genes can be classified as intergenic, intronic, and exonic. About 50% of genes of miRNAs are located within protein-encoding and non-protein-coding genes (host genes), mainly in introns and less frequently in exons. Genes of miRNAs can be transcribed from both independent promoters and the host gene promoter [7]. New miRNA genes are generated as a result of duplication of the existing miRNA genes (as it happens in the majority of cases) or de novo from the hairpin structures located within introns or intergenic regions [8]. De novo structures emerge by means of different mechanisms: 1) due to inverted duplication of the gene that will become a miRNA target in the future; 2) from transposons; 3) due to spontaneous evolution from random sequences [8].
The most common miRNA biogenesis pathway is referred to as canonical. In addition, other miRNA biogenesis pathways have been reported, which involve other proteins and in which one or more phases of canonical biogenesis are missing; they are referred to as non-canonical [9].
Canonical pathway of miRNA biogenesis
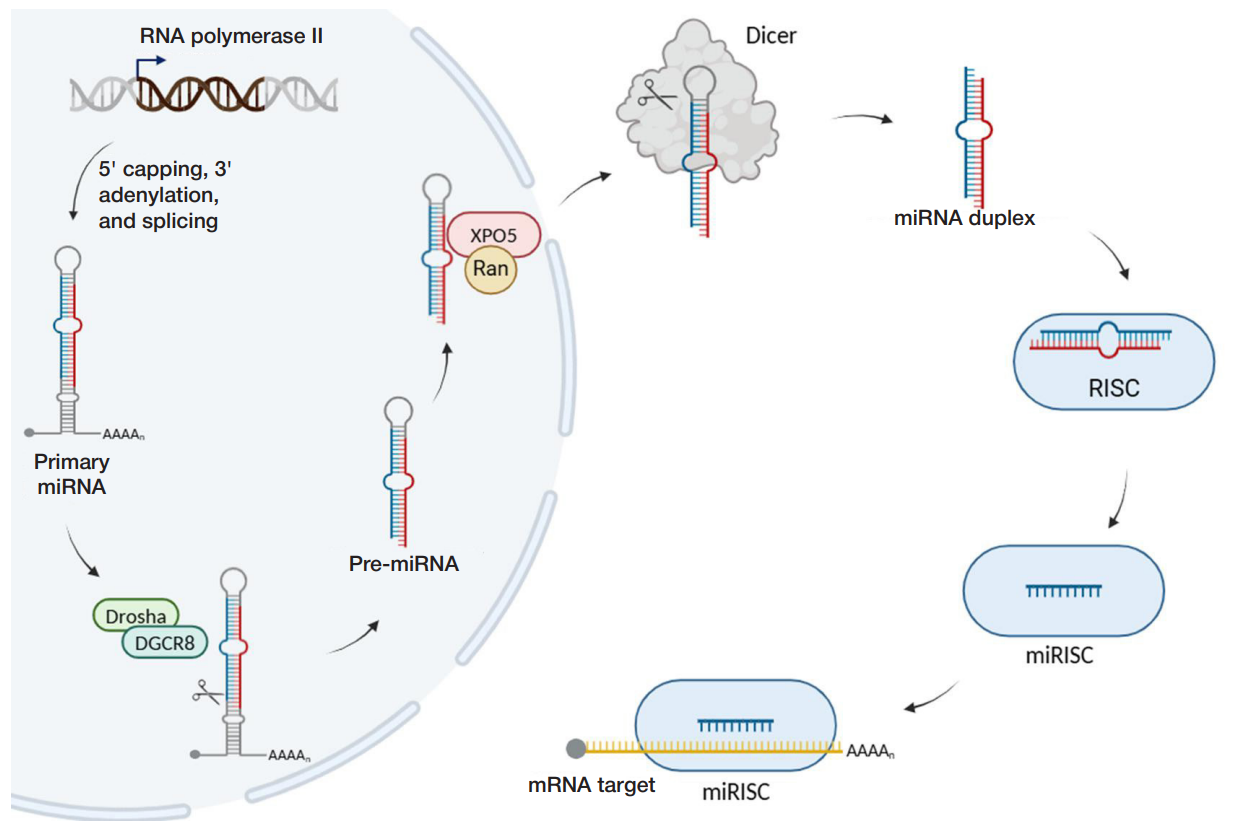
In animals, primary miRNA (pri-miRNA) is transcribed from the miRNA gene with the help of RNA polymerase II, and then undergoes 5' capping, 3' polyadenylation, and splicing (fig. 1).
Then a microprocessor complex consisting of ribonuclease III (Drosha) and the DGCR8 protein (DiGeorge Syndrome Critical Region 8 also referred to as Pasha — Partner of Drosha) cuts primary miRNA to pre-miRNA (precursor miRNA), 70—120 nucleotides in length. Pre-miRNA represents a hairpin consisting of the single-stranded region (terminal loop) and doublestranded stem with two nucleotides protruding at the 3' end. The protruding nucleotides are recognized by the exportin-5 (XPO5) protein, which transports pre-miRNA from the nucleus to the cytoplasm with the participation of the GTP-binding protein Ran. In the cytoplasm (fig. 1, on the right), ribonuclease III Dicer cuts the terminal loop out of pre-miRNA, which results in a duplex consisting of two mature miRNAs, 21–24 nucleotides in length [10]. Depending on the mature miRNA strand position in the pre-miRNA hairpin, their names are followed by suffixes -3p or -5p (for example, the hsa-miR-25-5p strand is located at the 5' end of the hsa-miR-25 pre-miRNA, while the hsamiR-25-3p strand is located at the 3' end). After cutting out the terminal loop, the miRNA duplex binds to the Argonaute (AGO) family protein being part of the RISC complex (RNA-induced silencing complex): thus the intermediate pre-RISC complex is formed. One of two strands of the RNA duplex dissociates and usually undergoes degradation, while the other remains loaded into the miRISC complex. The strand remaining loaded in this complex is referred to as guide strand, while the dissociated one is referred to as passenger strand [10]. This is followed by binding of the guide strand being part of the miRISC complex to the target mRNA.
In plants, the canonical pathway is slightly different from that of animals. Pri-miRNA is processed in the nucleus directly to the miRNA duplex by the Dicer protein homolog, DICERLIKE 1 (DCL1), which is responsible for both processing events essential for miRNA maturation [4]. In plants, the stem-loop precursor is longer and more variable. In contrast to bilateral animals, plant miRNAs undergo methylation by the HEN1 protein. Non-canonical pathways of miRNA biogenesis
Not all proteins of the canonical pathway are involved in processing in non-canonical (alternative) pathways of miRNA maturation. Drosha- and DGSR8-independent (microprocessorindependent), as well as Dicer-independent miRNA biogenesis pathways are distinguished. In the majority of Droshaindependent pathways, miRNA precursors represent products of processing of other RNAs (for example, small nuclear RNA or transfer RNA) and do not need to be hydrolyzed by the microprocessor complex [11]. In Dicer-independent processing, pri-miRNA is hydrolyzed by the microprocessor complex, but the resulting stem of the hairpin structure is too short for Dicer recognition. Therefore, the miRNA precursor is loaded directly onto the AGO protein, which incises one of the miRNA strands, and then the intermediate product obtained using the poly(A)specific ribonuclease (PARN) is truncated to produce the mature molecule [10].
Selection of functionally active miRNA strand
Selection of the guide strand between the -5p and -3p strands is associated with thermodynamic instability of the duplex and miRNA nucleotide composition [12]. The 5' end of miRNA duplex is incorporated in specific pocket formed by the MID and PIWI domains of the AGO protein. These domains are sensitive to the nucleotide composition of the duplex and bind uracil twice stronger, than adenine, and 30 times stronger, than cytosine or guanine, i.e. preference is given to the strand that is uracil-rich at the 5' end. AGO is also likely to load the strand showing lower relative thermodynamic stability. It is believed that the first four nucleotides at each end of the duplex are responsible for thermodynamic stability, and the difference in one additional hydrogen bond at one end is enough to affect the choice of the guide and passenger strands. Usually, 5' ends of the degrading passenger strands are pyrimidine-rich, while those of guide strands are purine-rich [12].
Target mRNA recognition and binding by the miRNA molecule
The miRISC complex bound to the target mRNA interacts with the GW182 protein (182 kDa glycine-tryptophan repeatcontaining protein). GW182 functions as a molecular scaffold for linking of the AGO protein being part of the miRISC complex and the downstream effector complexes involved in the miRNA-mediated translational repression of the target mRNA [7].
To facilitate target mRNA recognition and binding, AGO protein ensures spatial positioning of the miRNA guide strand. The miRNA guide strand region between nucleotides 2 and 7 at the 5′ end is referred to as seed region; it is responsible for target mRNA binding and is of crucial importance for its recognition [13]. Nucleotides 8 and 13–16 of the miRNA sequence are also involved in recognition [10]. In the mRNA target molecule, the miRISC binding site is usually in the 3'-untranslated region (3'-UTR). Rather rare non-canonical binding sites in the coding and 5'-UTR regions of mRNA have been reported, during interaction with which miRNAs also suppress mRNA translation. In some cases, miRNAs can interact with promoter regions of genes, thereby ensuring activation of their transcription [14].
miRNA-mediated gene silencing mechanisms
As stated above, recognition of the target mRNA by the miRNA molecule usually leads to suppression of translation of this mRNA. In the case of complete complementarity observed mainly in plants, the target mRNA is cleaved within miRISC by the AGO protein with RNAse Н endonuclease activity, in the region complementary to nucleotides 10 and 11 of the miRNA guide strand (endonuclease cleavage of mRNA) [4, 15]. In the majority of animal cells, binding sites within mRNA are not fully complementary to the miRNA seed region. In this case, the miRISC-mediated translation repression and destabilization of mRNA with subsequent degradation are initiated [7].
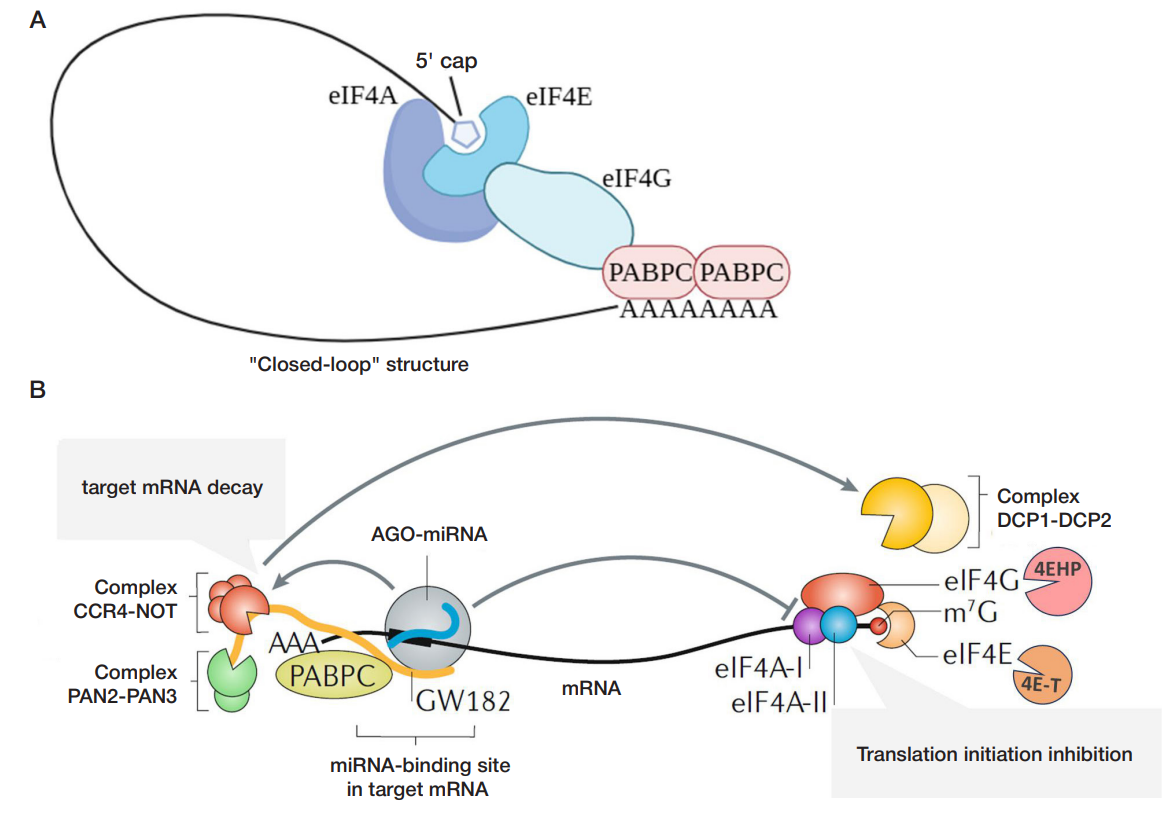
Despite the fact that the exact mechanisms underlying these processes in mammals remain the matter of debate, it is generally accepted that initially translation repression occurs, followed by destabilization and decay of the target mRNA [16]. Several mechanisms underlying repression of translation have been reported, among which inhibition of the formation of a “closed-loop” structure of the target mRNA (fig. 2A). The PABPC protein bound to the 3′ poly(A) tail of the target mRNA stabilizes interaction between the 5′ cap and the eukaryotic translation initiation factor 4F complex consisting of eIF4A, eIF4G and eIF4E through binding to eIF4G. The mRNA ends are brought close together, and the “closed-loop” structure is formed, and thus facilitates translation initiation and ribosome recycling.
The AGO protein being part of the miRISC complex causes dissociation of the PABPС protein, which leads to aberrant “closed-loop” formation and inhibition of translation initiation. Other mechanisms include inhibition of mRNA translation via prevention of the eukaryotic translation initiation factor 4F complex assembly. These include attraction of the 4E-T and 4EHP translation inhibitors by the miRISC complex and subsequent dissociation of eIF4E and eIF4G or dissociation of the eIF4A subunit (fig. 2B, on the right). The mechanisms are not mutually exclusive; they can work at different rates at the same time. Predominance of certain mechanism depends on the nature of target mRNA and the cell type [16]. Repression of translation is followed by destabilization of the target mRNA, including deadenylation and decapping with subsequent exonucleolytic degradation of mRNA (miRNAmediated target mRNA decay) (fig. 2B, on the left). First, proteins of the AGO family recruit the GW182 protein, which interacts with the РАВРC poly(A)-binding protein and causes its dissociation from the mRNA poly(A) tail. This increases accessibility of the mRNA poly(A) tail for the CCR4-NOT and PAN2-PAN3 deadenylases, which are also recruited by the GW182 protein. As a result, mRNA deadenylation occurs. Furthermore, the CCR4-NOT complex recruits the DCP1 and DCP2 proteins dacapping target mRNA targets, which makes mRNA accessible for degradation by the XRN1 exonuclease [16]. miRNA is characterized by redundancy: expression of one mRNA can be regulated by various miRNAs interacting with its different binding sites. Furthermore, miRNA is pleiotropic: one miRNA can interact with various target mRNAs. In animals, one miRNA has much more targets, than in plants, due to frequently occurring incomplete complementarity [4]. The combination of such properties results in generation of the complex miRNAmediated regulatory network.
miRNA localization in the cell
Previously it was assumed that miRNA performs its function in the cytoplasm only. However, recent studies have shown that the cytoplasmic miRISC complex can be imported to the nucleus [18]. The nuclear miRISC complex is either similar to cytoplasmic one in components, or can exist as a separate AGO-miRNA complex. The nuclear miRISC binds to complementary nuclear transcripts or, if there are none, is exported to the cytoplasm again. Accumulation of miRNA in the cytoplasm or nucleus is partially determined by localization of its target mRNAs. In the nucleus, miRNA can perform several functions: 1) gene expression regulation via binding to their promoter; 2) binding and suppression of function of the long non-coding RNAs; 3) disruption of miRNA biogenesis via binding to pri-miRNA; 4) fine-tuning of target mRNA expression via miRNA retention in the nucleus [19].
It is well known that miRNAs can be released from the cell, to be carried by blood, and to affect other cells of the body, mediating intercellular communication. Outside the cell, miRNAs can be contained in the membrane vesicles (exosomes, microvesicles), as well as in the apoptotic bodies formed when the cells die. In the extracellular environment, the majority of miRNAs (90%) are carried by AGO proteins, the other 10% circulate as complexes with high-density lipoproteins [20].
miRNA involvement in physiological and pathological processes in the cell
miRNAs regulate almost all processes in the cells: metabolism, cell cycle progression, proliferation, differentiation, and apoptosis, so disruption of their regulatory properties can lead to various diseases [21]. Actually, alterations of miRNA levels in humans are often observed in a broad spectrum of pathologies: tumors, cardiovascular, neurological and autoimmune diseases, inflammation, gastrointestinal tract and skeletal muscle diseases [22]. The role of miRNA has been extensively studied in the context of various disorders since the moment of their discovery, and to date, information has been accumulated about what levels of microRNAs most frequently change in affected tissues in certain conditions, which miRNAs have protective effects and which, on the contrary, contribute to developing pathological phenotype. miRNAs are examined both directly in the patients’ tissue specimens and in in vivo, in vitro experiments, as well as in silico, using the latter approach to construct preliminary hypotheses. Among all patients’ biomaterials, biopsy specimens of affected tissues are best suited for investigation of the role of miRNAs in etiopathogenesis, but obtaining them is associated with invasive interventions, so it is usually hardly accessible. In this regard, circulating miRNAs most often become the research object. These are miRNAs that are released into the extracellular space due to tissue damage, apoptosis, necrosis or intense secretion and circulate in the body [7]. Such miRNAs are found in blood serum and plasma, urine, tears, saliva, seminal, peritoneal and cerebrospinal fluids, breast milk and bronchoalveolar lavage [7]. They can regulate the target cell activity, thereby acting as intercellular signaling molecules. For example, endothelium-released miRNAs regulate activity of vascular smooth muscle cells, while oncoogenic miRNAs in exosomes increase invasiveness of breast cancer cells [23, 24].
Circulating miRNAs are stable at high pH and temperature and are resistant to degradation, so they seem to be attractive as candidate biomarkers. Actually, there are studies, in which such circulating molecules are used for differential diagnosis, early diagnosis, prediction of the disease course, disease severity assessment, or control over response to therapy [25]. Since miRNAs can regulate translation of several mRNA targets, some authors propose to evaluate diagnostic value of not only one specific miRNA, but the set of miRNAs (signitures) that would be more specific for the disease [26]. For example, assessment of urinary levels of the set of three miRNAs in individuals with lupus nephritis makes it possible to reveal early renal fibrosis and predict the development of kidney failure [27].
The use of miRNAs for therapeutic purposes is based either on replenishing the levels of essential miRNAs via administration of appropriate synthetic analogues (miRNA mimetics), or on inhibition of target miRNAs via administration of antisense miRNA or sponge RNAs (also referred to as competing endogenous RNAs such as pseudogene transcripts, long non-coding RNAs, circular RNAs and mRNAs, which bind the pool of miRNAs, competing with their targets). Thus, miR-34a suppresses expression of more than 30 oncogenes involved in the tumor cell evasion from the immune system. Since expression of this miRNA is often decreased in various malignant tumors, a synthetic miR-34a mimetic, MRX34, has been developed for treatment of malignant tumors of the skin, lung, kidney, and liver [28]. Currently, a number of biopharmaceutical companies (Santaris Pharma, Roche Pharmaceuticals, Regulus therapeutics, Mirna Therapeutics Inc., miRagen Therapeutics and EnGeneIC) have programs focused on using miRNAs for therapy of tumors. Despite the fact that several clinical trials of miRNAs as potential drugs were terminated due to serious side effects and toxicity, studies of the use of miRNAs in therapeutic practice continue and inspire great hope [29].
CONCLUSION
Thirty years since miRNA discovery is a long time, during which the number of miRNA studies has been increasing steadily. The data accumulated during this period have both expanded our knowledge about the mechanisms underlying gene activity regulation and opened up new horizons in understanding their role in body’s physiological and disease processes. In many disorders miRNAs can serve as reliable biomarkers, and in a number of cases miRNA activity modulation is used for treatment of various diseases. However, comprehensive analysis of certain miRNA involvement in cellular processes or pathogenesis of certain diseases is beyond the scope of this review. We have tried to make the reader familiar with the broad range of capabilities of these small RNAs, as well as with the prospects for their further study and use.
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993; 75 (5): 843–54. DOI:10.1016/0092-8674(93)90529-Y
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993; 75 (5): 855–62. DOI: 10.1016/0092-8674(93)90530-4.
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000; 403 (6772): 901–06. DOI: 10.1038/35002607.
- Moran Y, Agron M, Praher D, Technau U. The evolutionary origin of plant and animal microRNAs. Nat Ecol Evol. 2017; 1 (3): 27. DOI: 10.1038/s41559-016-0027.
- Dexheimer PJ, Cochella L. MicroRNAs: From Mechanism to Organism. Front Cell Dev Biol. 2020; 8: 409. DOI: 10.3389/fcell.2020.00409.
- Bhaskaran M, Mohan M. MicroRNAs: History, Biogenesis, and Their Evolving Role in Animal Development and Disease. Vet pathol. 2013; 51 (4): 759. DOI: 10.1177/0300985813502820.
- O’Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne). 2018; 9: 402. DOI: 10.3389/fendo.2018.00402.
- Liu N, Okamura K, Tyler DM, Phillips MD, Chung WJ, Lai EC. The evolution and functional diversification of animal microRNA genes. Cell Res. 2008; 18 (10): 985. DOI: 10.1038/cr.2008.278.
- Abdelfattah AM, Park C, Choi MY. Update on non-canonical microRNAs. Biomol Concepts. 2014; 5 (4): 275. DOI: 10.1515/bmc-2014-0012.
- Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019; 20 (1): 5–20. DOI: 10.1038/s41580-018-0059-1.
- Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011; 43 (6): 892–903. DOI: 10.1016/j.molcel.2011.07.024.
- Medley JC, Panzade G, Zinovyeva AY. microRNA strand selection: Unwinding the rules. Wiley Interdiscip Rev RNA. 2021; 12 (3): e1627. DOI: 10.1002/wrna.1627.
- Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015; 4: e05005. DOI: 10.7554/eLife.05005.
- Broughton JP, Lovci MT, Huang JL, Yeo GW, Pasquinelli AE. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol Diseases. Avicenna J Med Biotechnol. 2010; 2 (4): 161.
- Pratt AJ, MacRae IJ. The RNA-induced Silencing Complex: A Versatile Gene-silencing Machine. J Biol Chem. 2009; 284 (27): 17897. DOI:10.1074/jbc.R900012200.
- Iwakawa HO, Tomari Y. Life of RISC: Formation, action, and degradation of RNA-induced silencing complex. Mol Cell. 2022; 82 (1): 30–43. DOI: 10.1016/j.molcel.2021.11.026.
- Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019; 20 (1): 21–37. DOI: 10.1038/s41580-018-0045-7.
- Wong JJL, Ritchie W, Gao D, Lau KA, Gonzalez M, Choudhary A, et al. Identification of nuclear-enriched miRNAs during mouse granulopoiesis. J Hematol Oncol. 2014; 7: 42. DOI: 10.1186/1756-8722-7-42.
- Hu X, Yin G, Zhang Y, Zhu L, Huang H, Lv K. Recent advances in the functional explorations of nuclear microRNAs. Front Immunol. 2023; 14: 1097491. DOI: 10.3389/fimmu.2023.1097491.
- Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012; 110 (3): 483–95. DOI: 10.1161/CIRCRESAHA.111.247452.
- Ardekani AM, Naeini MM. The Role of MicroRNAs in Human Diseases. Avicenna J Med Biotechnol. 2010; 2 (4): 161.
- Paul P, Chakraborty A, Sarkar D, Langthasa M, Rahman M, Bari M, et al. Interplay between miRNAs and human diseases. J Cell Physiol. 2018; 233 (3): 2007–18. DOI: 10.1002/jcp.25854.
- Zhu JJ, Liu YF, Zhang YP, Zhao CR, Yao WJ, Li YS, et al. VAMP3 and SNAP23 mediate the disturbed flow-induced endothelial microRNA secretion and smooth muscle hyperplasia. Proc Natl Acad Sci U S A. 2017; 114 (31): 8271. DOI: 10.1073/pnas.1700561114.
- Yang M, Chen J, Su F, Yu B, Su F, Lin L, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011; 10: 117. DOI: 10.1186/1476-4598-10-117.
- Ho PTB, Clark IM, Le LTT. MicroRNA-Based Diagnosis and Therapy. Int J Mol Sci. 2022; 23 (13): 7167. DOI: 10.3390/ijms23137167.
- Backes C, Meese E, Keller A. Specific miRNA Disease Biomarkers in Blood, Serum and Plasma: Challenges and Prospects. Mol Diagn Ther. 2016; 20 (6): 509–18. DOI: 10.1007/s40291-016-0221-4.
- Solé C, Moliné T, Vidal M, Ordi-Ros J, Cortés-Hernández J. An Exosomal Urinary miRNA Signature for Early Diagnosis of Renal Fibrosis in Lupus Nephritis. Cells. 2019; 8 (8): 773. DOI: 10.3390/cells8080773.
- Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, et al. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest new drugs. 2016; 35 (2): 180. DOI: 10.1007/s10637-016-0407-y.
- Seyhan AA. Trials and Tribulations of MicroRNA Therapeutics. Int J Mol Sci. 2024; 25 (3): 1469. DOI: 10.3390/ijms25031469.