This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (CC BY).
ORIGINAL RESEARCH
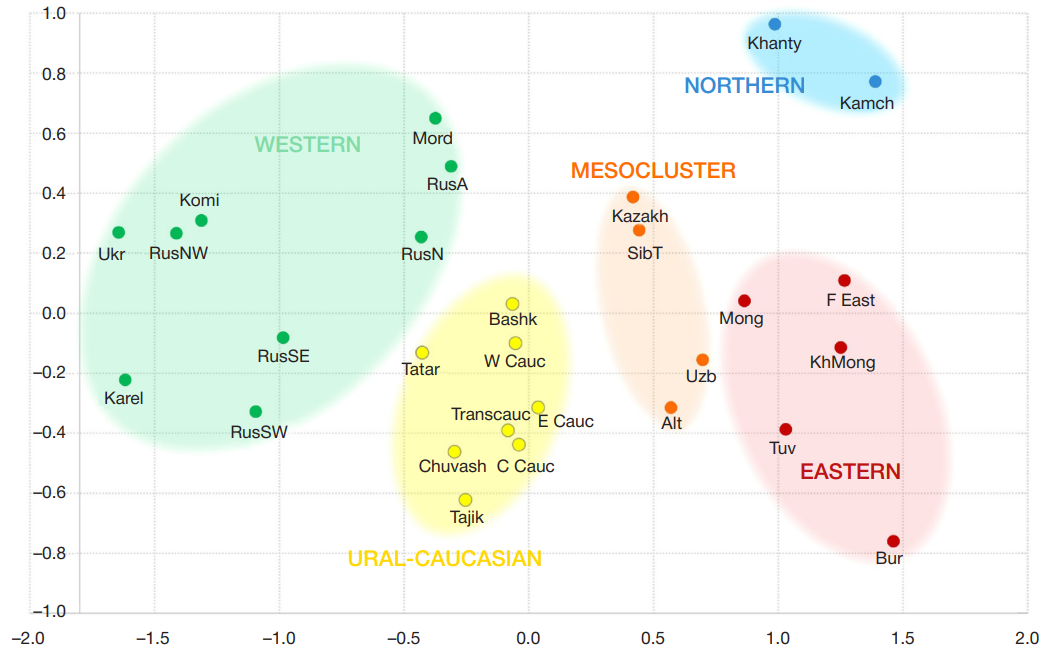
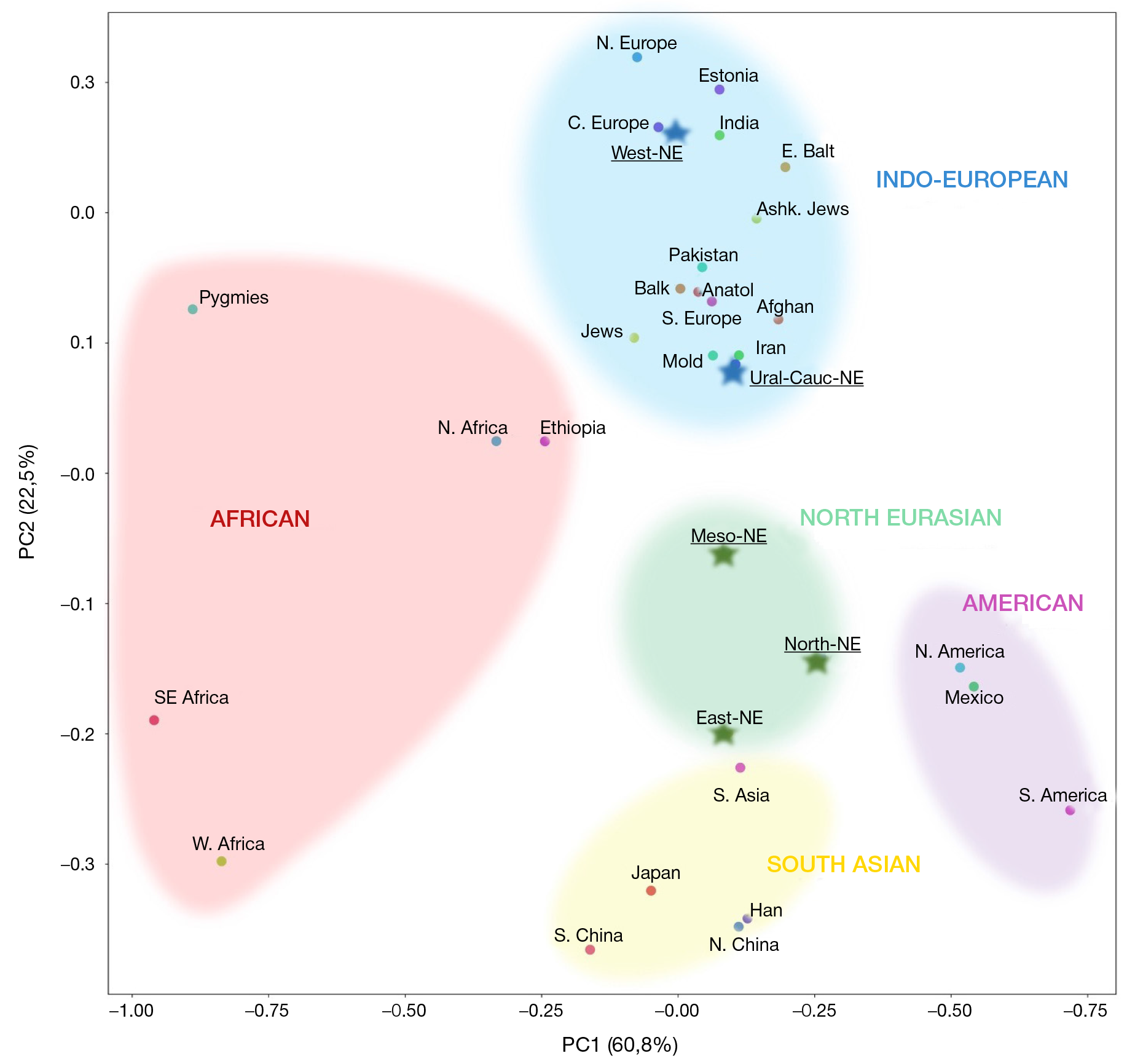
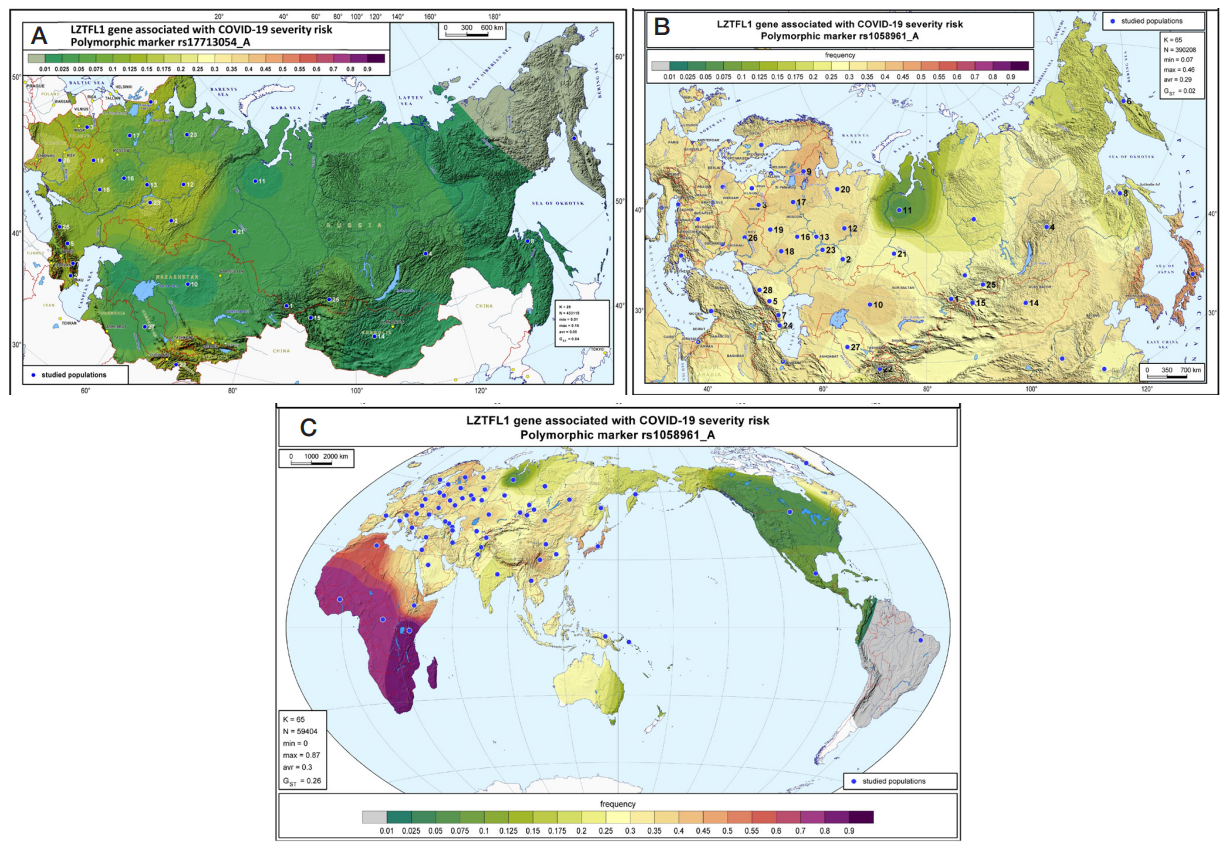
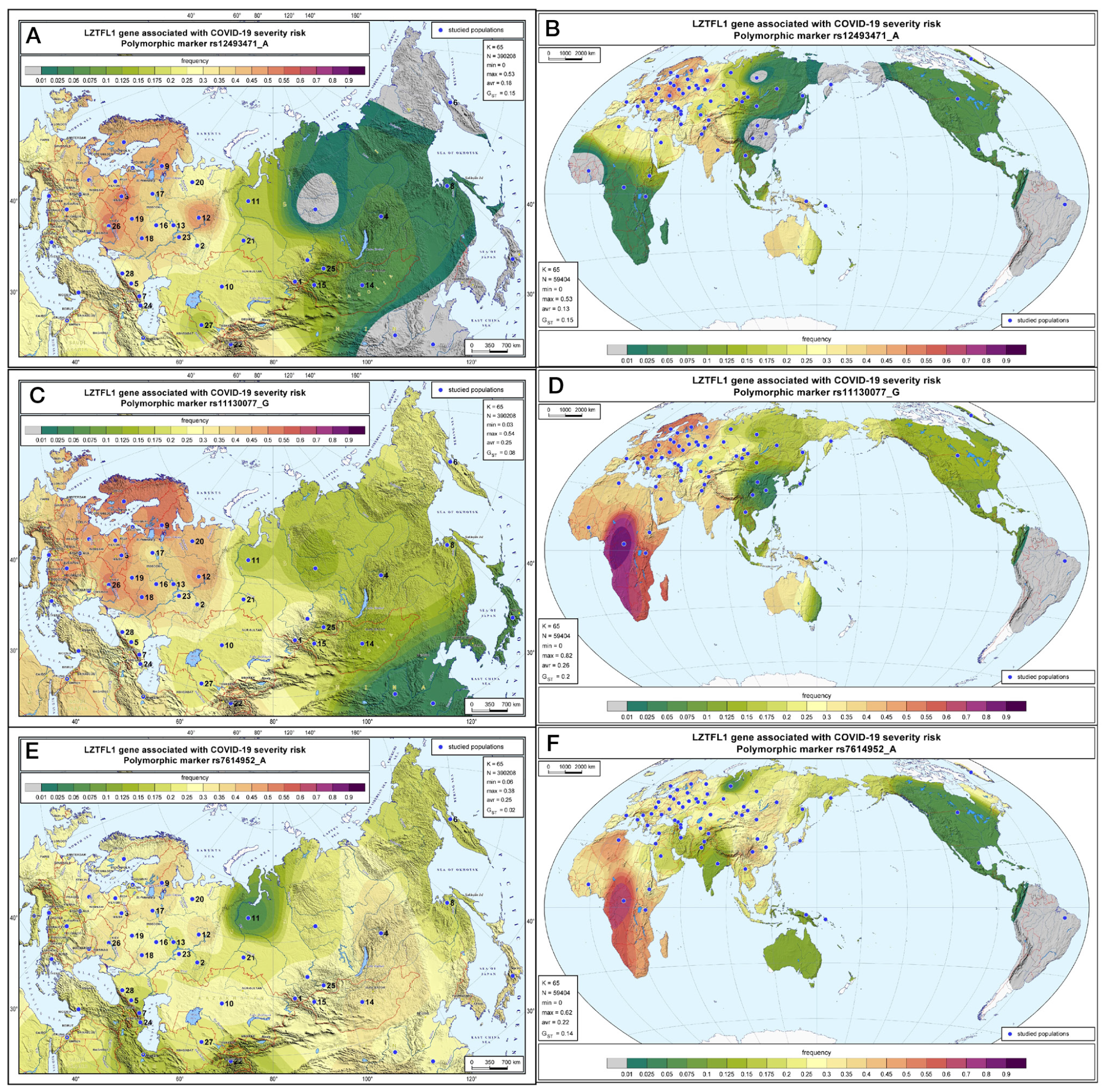
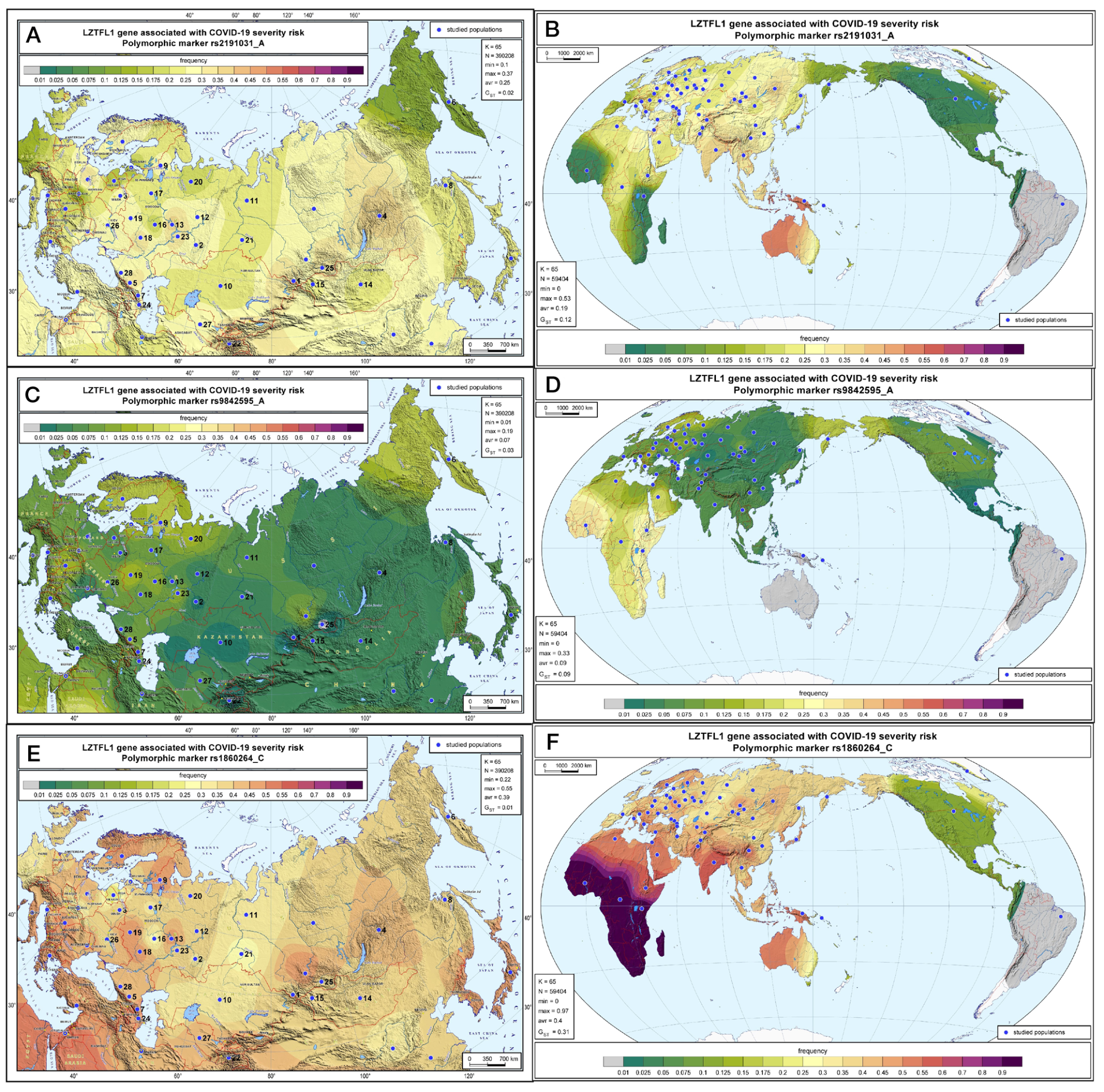
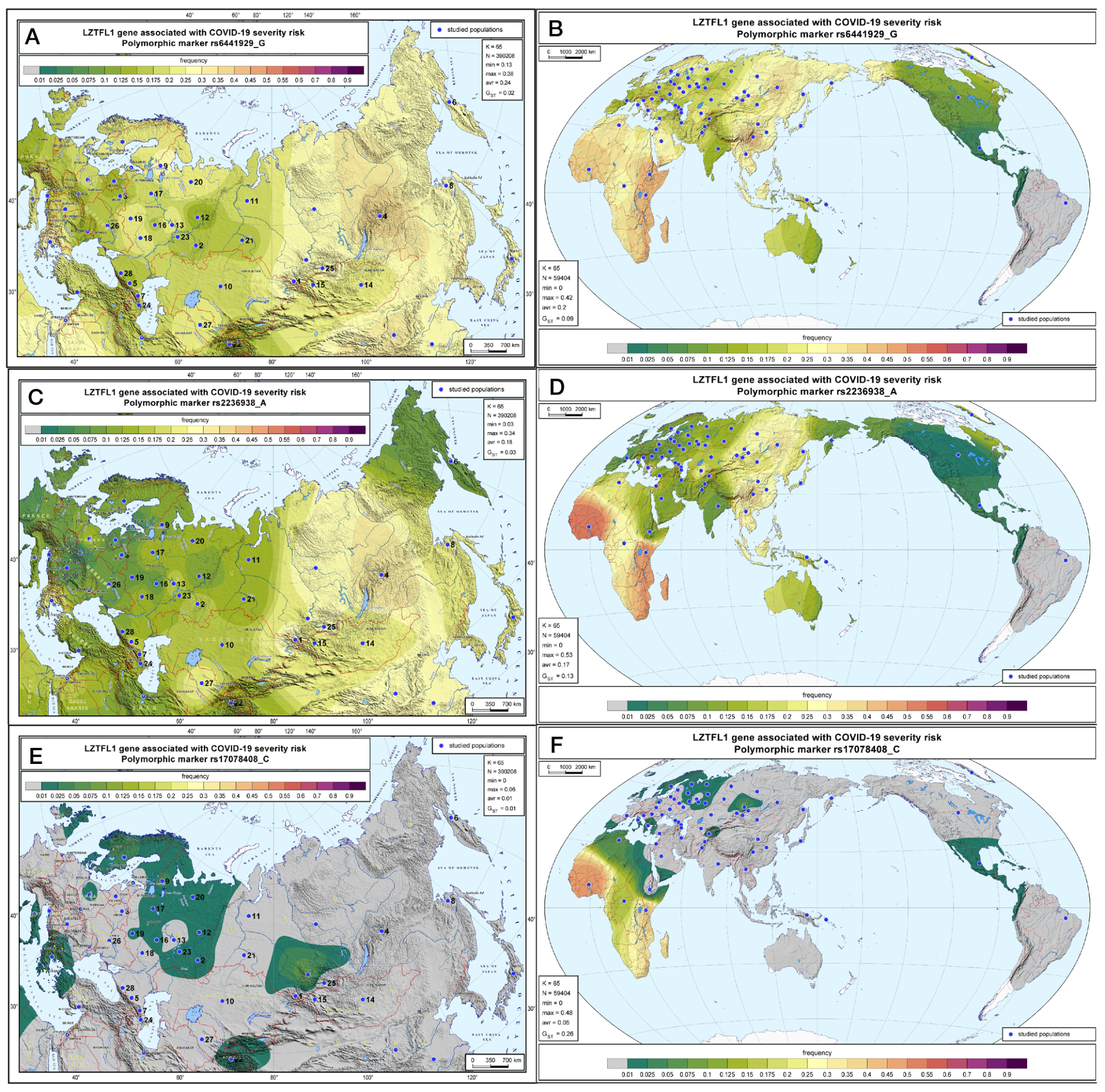
Geographic distribution of the LZTFL1 SNP markers associated with severe COVID-19 in Russia and worldwide
1 Research Centre for Medical Genetics, Moscow, Russia
2 Autonomous non-profit organization “Biobank of North Eurasia”, Moscow, Russia
3 Lomonosov Moscow State University, Moscow, Russia
4 Russian Medical Academy of Continuous Professional Education, Ministry of Healthcare of the Russian Federation, Moscow, Russia
Correspondence should be addressed: Elena V. Balanovska
Moskvorechye, 1, 115522, Moscow, Russia; ur.liam@aksvonalab
Funding: the study was supported by the Russian Science Foundation grant № 21-14-00363 (bioinformatics analysis, cartographic analysis) and performed within the State Assignment of the Ministry of Science and Higher Education of the Russian Federation for the Research Centre for Medical Genetics (statistical analysis, data interpretation, manuscript writing).
Acknowledgements: the authors express their gratitude to all members of the expedition survey of the North Eurasian indigenous population (sample donors) and the autonomous non-profit organization “Biobank of North Eurasia” for access to DNA collections, and Olkova MV, for her participation in gathering information on the gene variants associated with the COVID-19 severity.
Author contribution: Balanovska EV — data analysis, manuscript writing, research management; Gorin IO, Petrushenko VS — bioinformatics analysis; Agdzhoyan AT, Chernevsky DK, Pylev VYu — statistical analysis; Temirbulatov II — explanation of pharmacogenetic approaches; Koshel SM — cartographic analysis.
Compliance with ethical standards: the study was approved by the Ethics Commitee of the Research Centre for Medical Genetics (protocol № 1 of 29 June 2020); all subjects submitted the informed consent to study participation.