This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (CC BY).
ORIGINAL RESEARCH
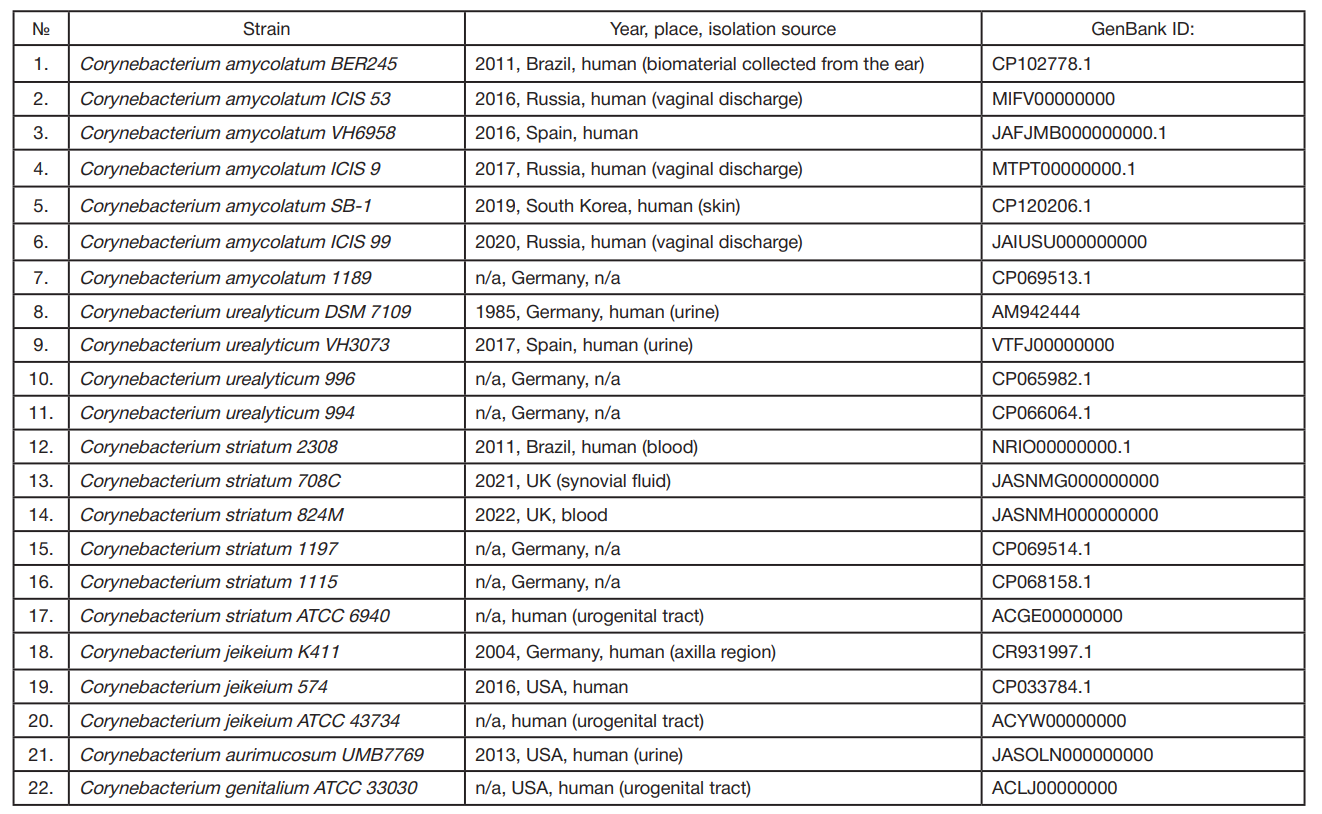
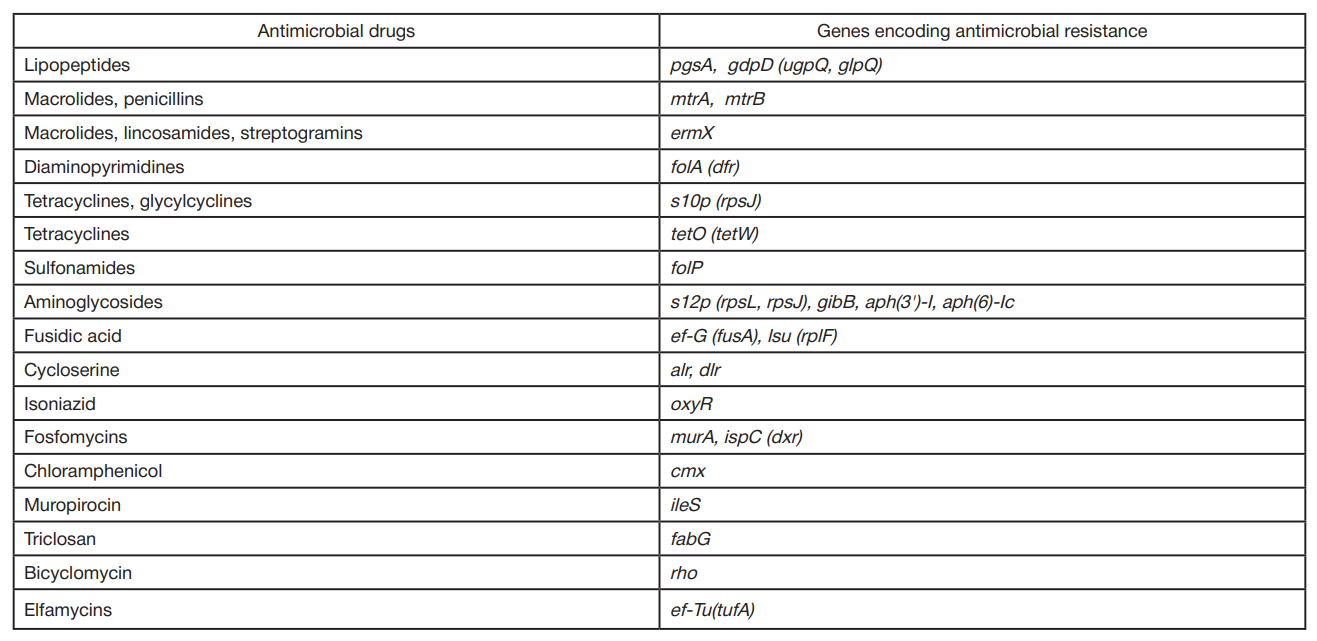
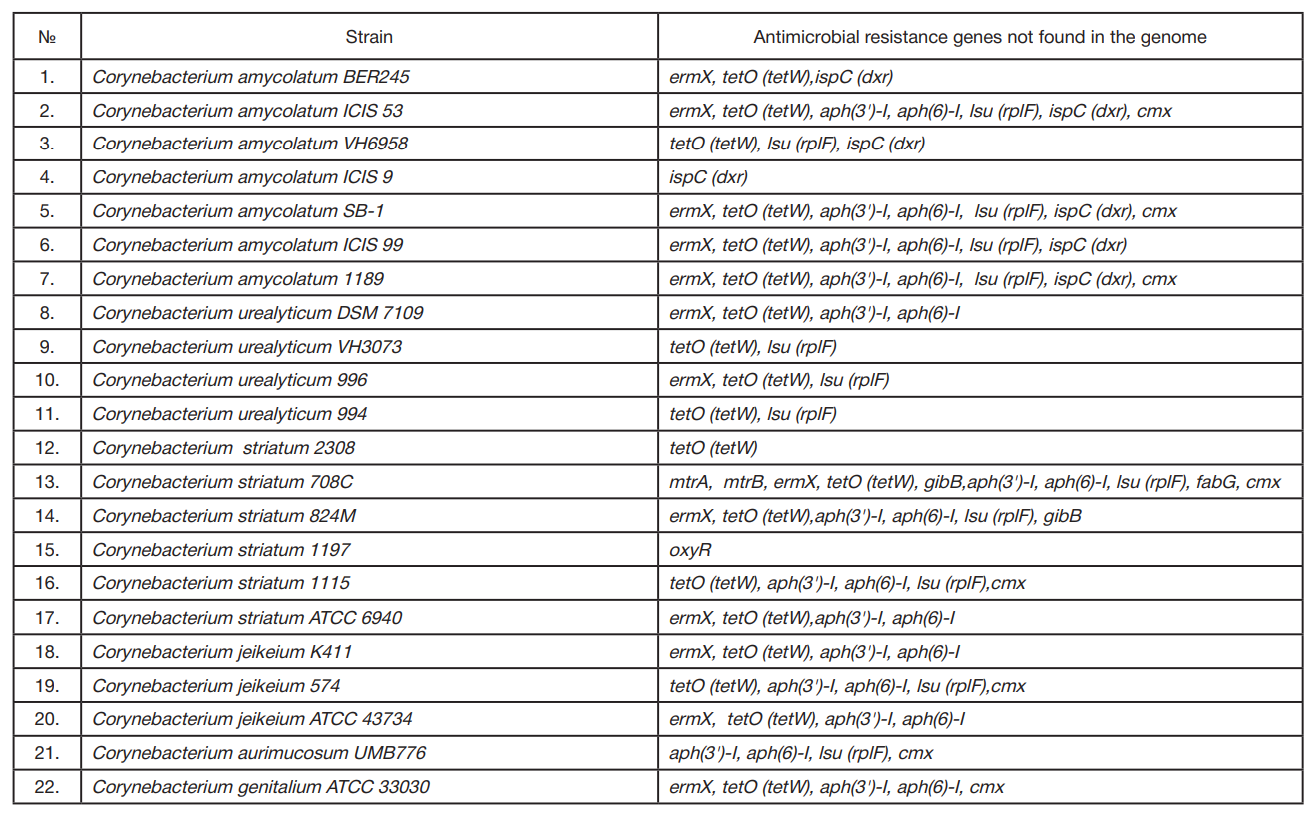
Comparative bioinformatics analysis of antimicrobial resistance gene pool in the genomes of representatives of genus Corynebacterium
Razumovsky Saratov State Medical University, Saratov, Russia
Correspondence should be addressed: Tatiana А. Kulshan
B. Kazachiya, 112, Saratov, 410012, Russia; ur.xednay@nahsluk.anajtat
Author contribution: Kulshan TА — study planning, literature review, dealing with molecular genetic data (selection of genomes, genome annotation, comparative analysis of gyrA amino acid sequences), data analysis, manuscript writing; Bugaeva IO — study planning, data analysis, interpretation of findings, manuscript writing; Soboleva EF — literature review, data analysis, manuscript writing; Allyanova MS — literature review, analysis of the antimicrobial resistance gene pool in the genomes of corynebacterial strains, dealing with the PATRIC online service; Popov DA — literature review, search for gyrA amino acid sequences in the genomes of corynebacterial strains, comparative analysis of amino acid sequences; Shvidenko IG — advising during manuscript writing, data analysis.