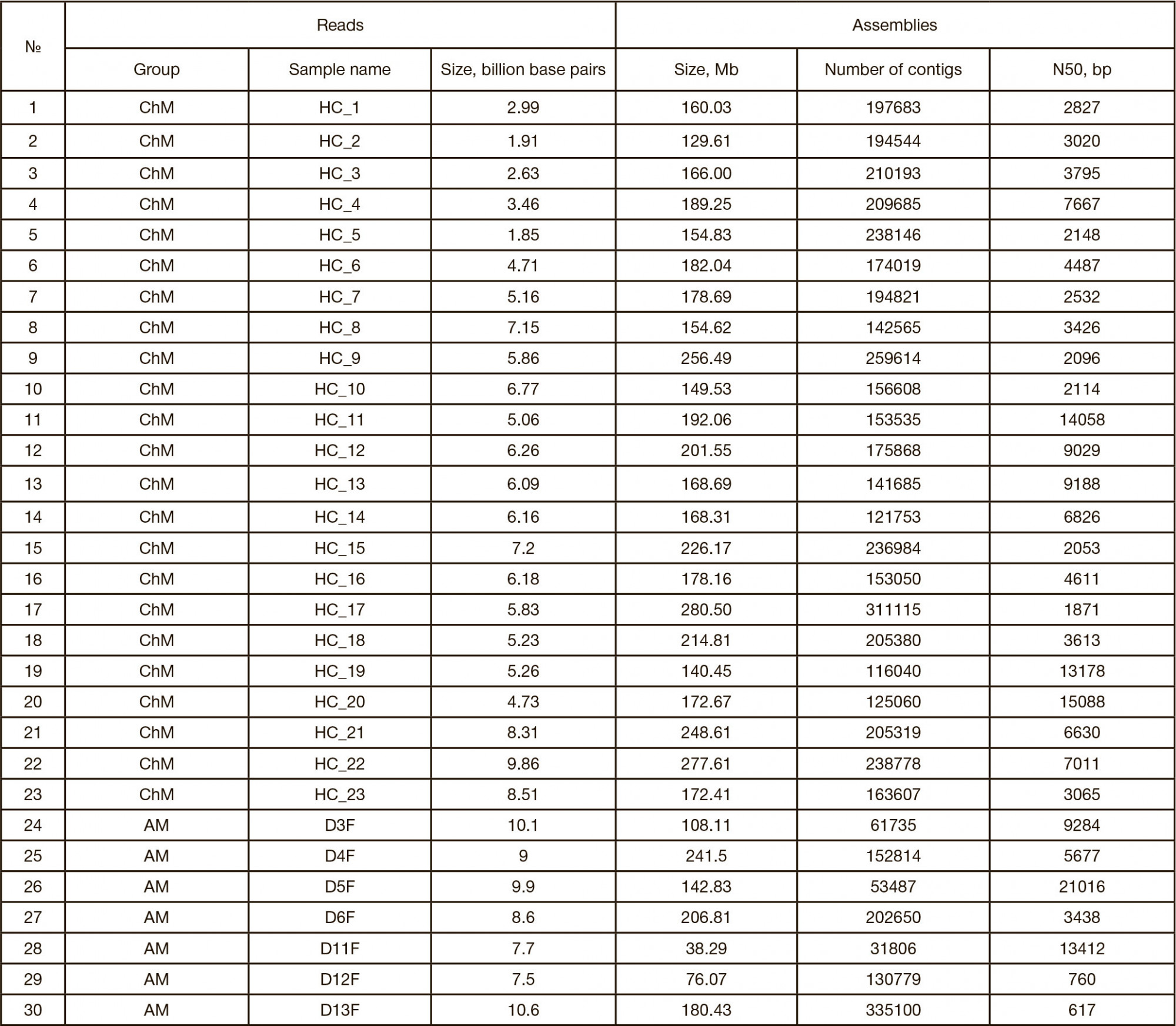
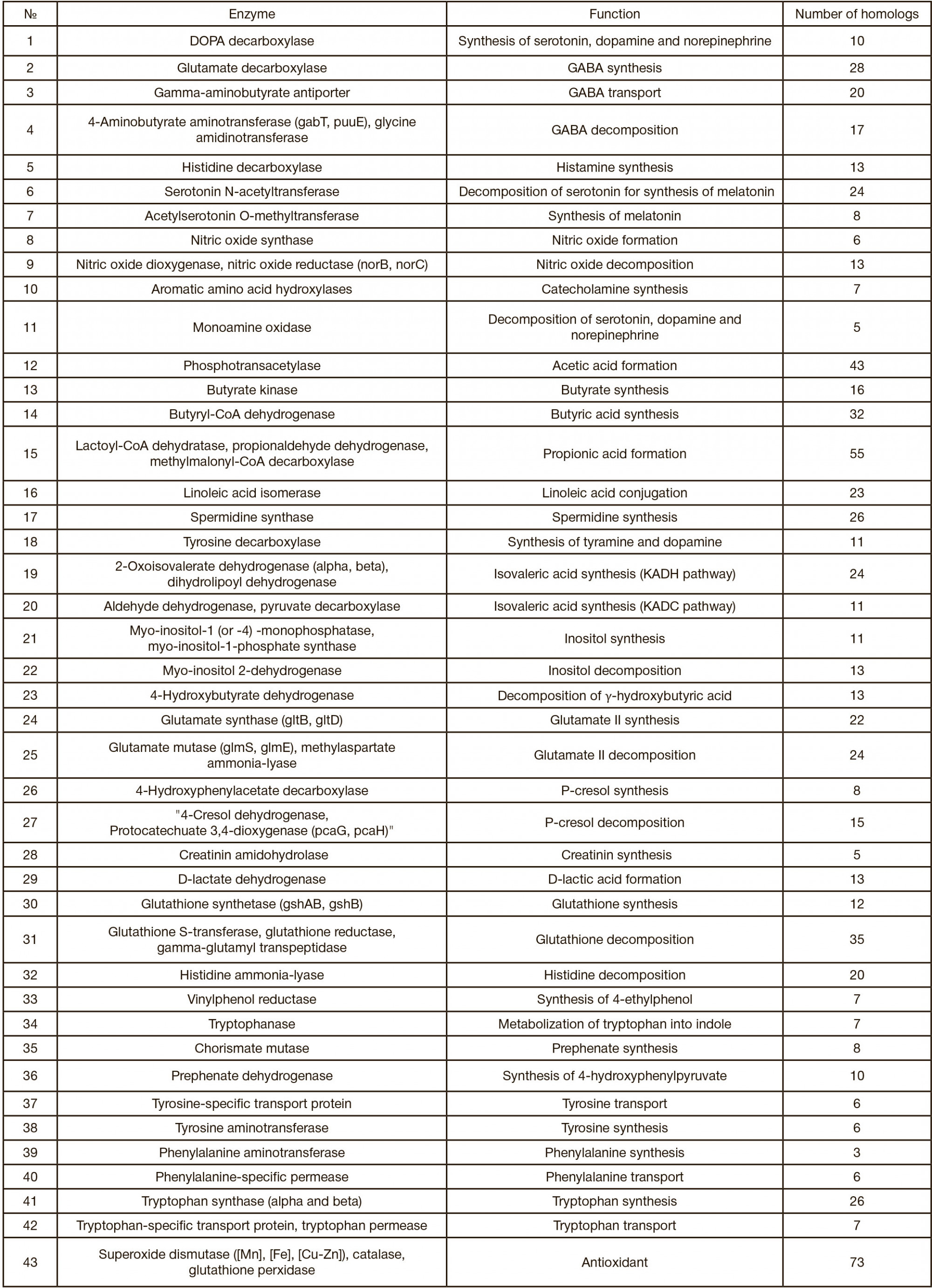
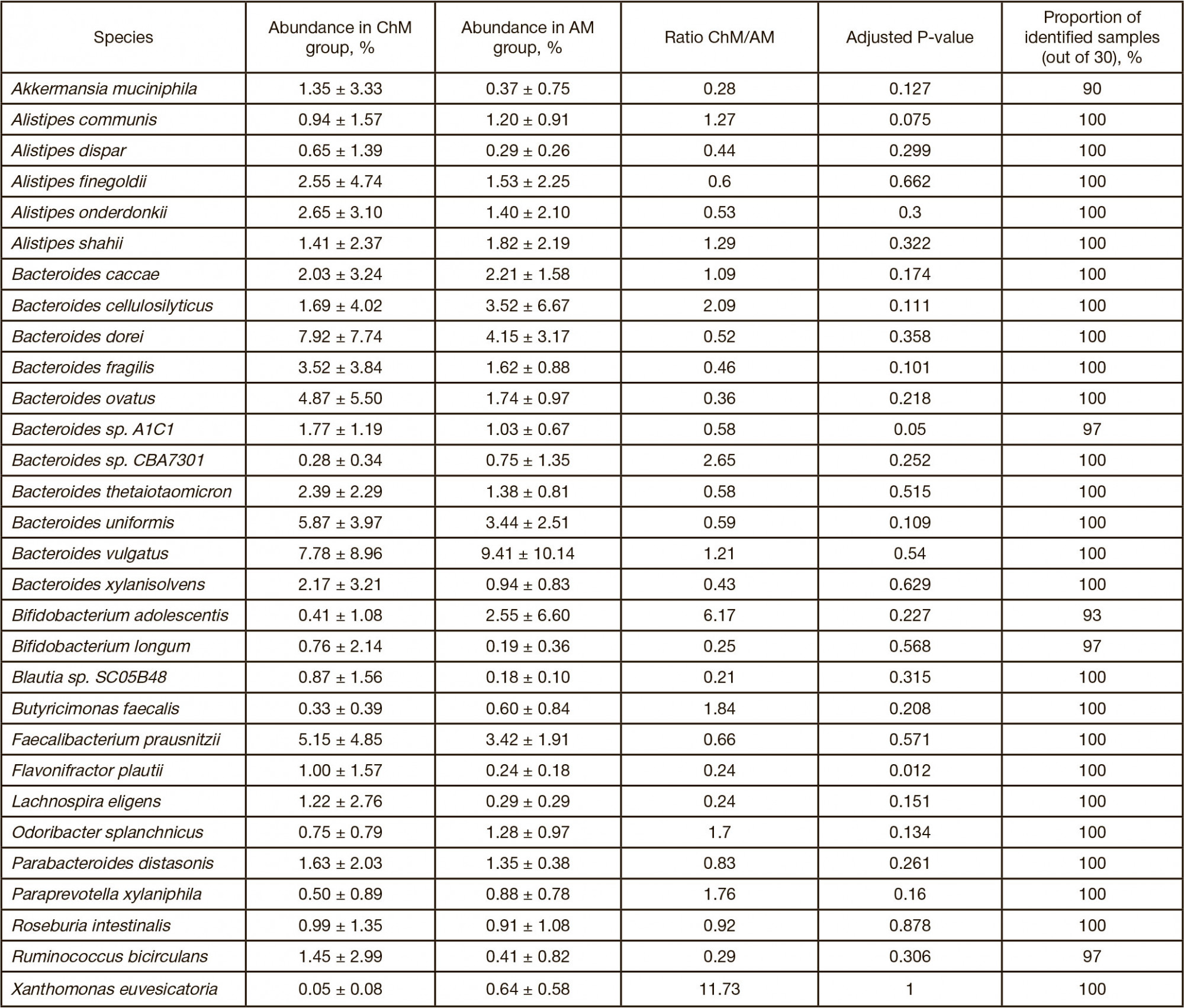
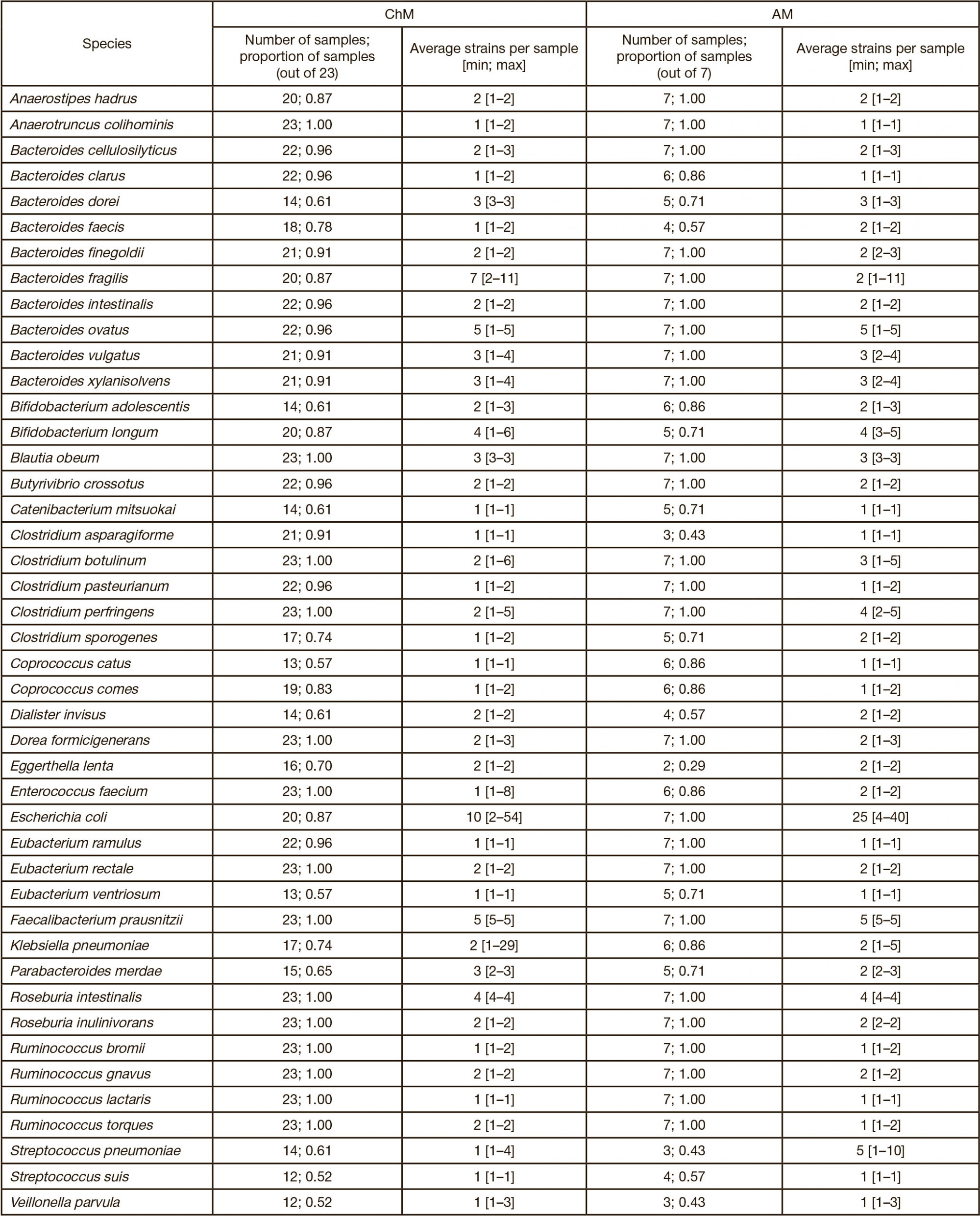
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (CC BY).
ORIGINAL RESEARCH
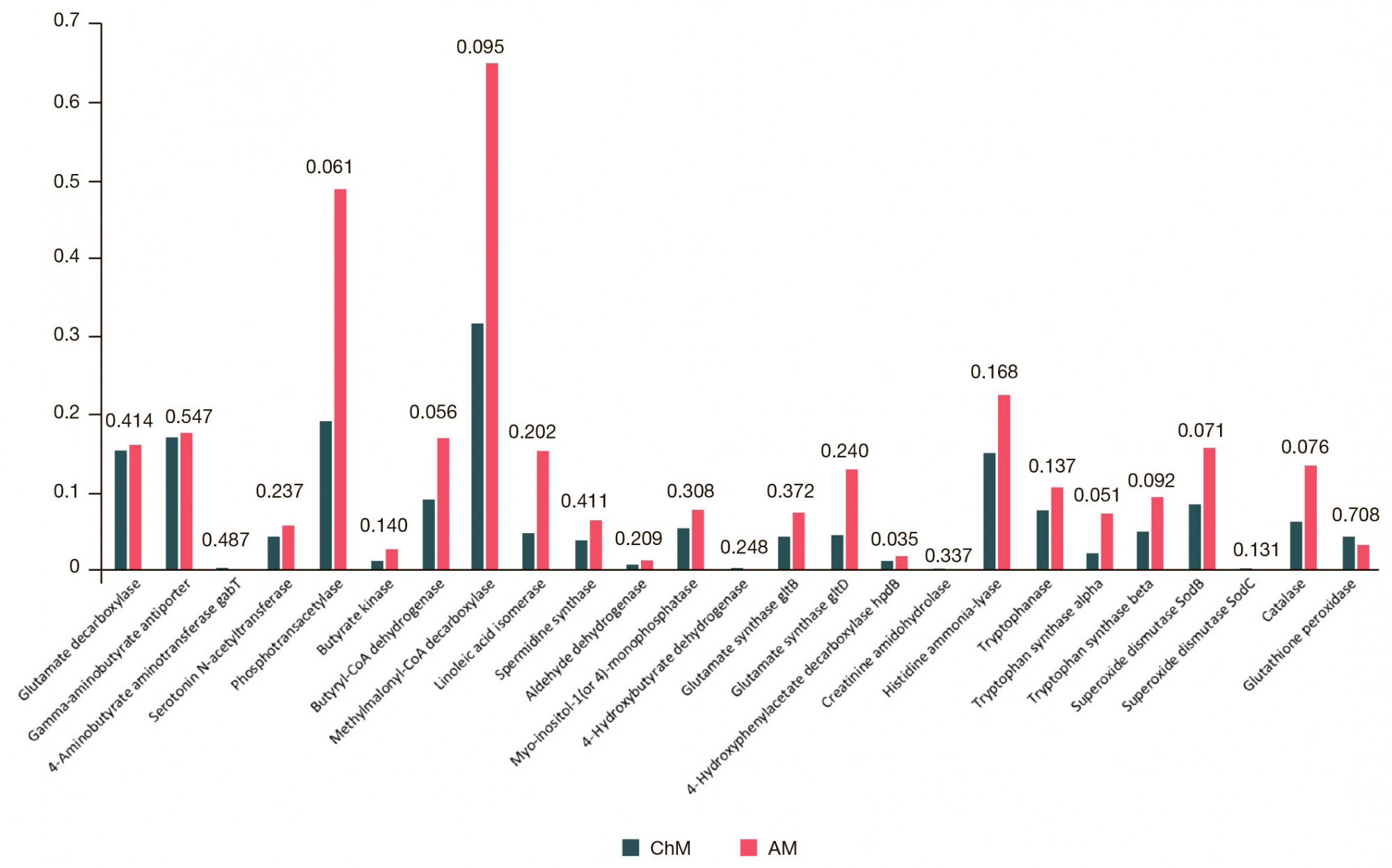
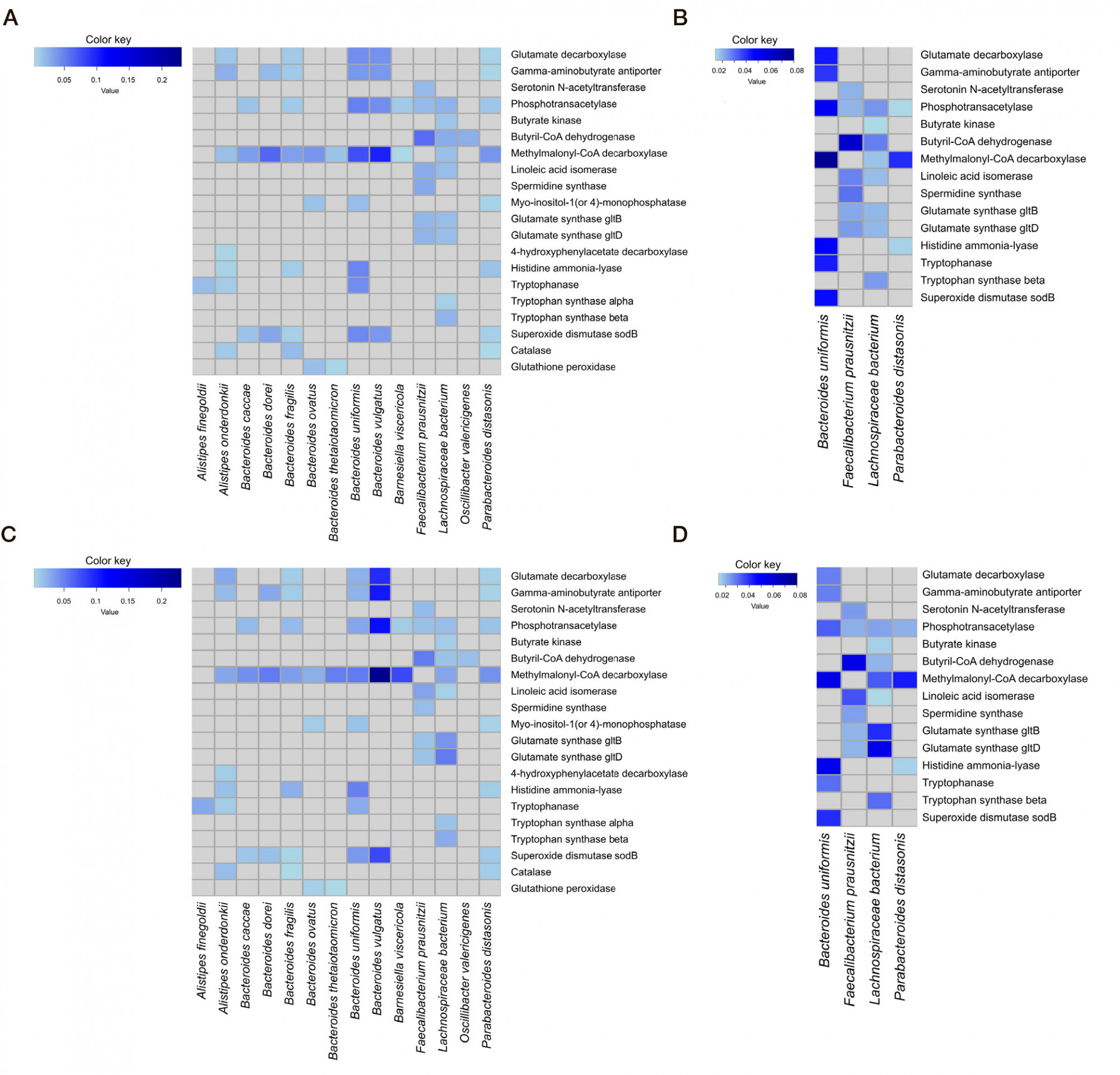
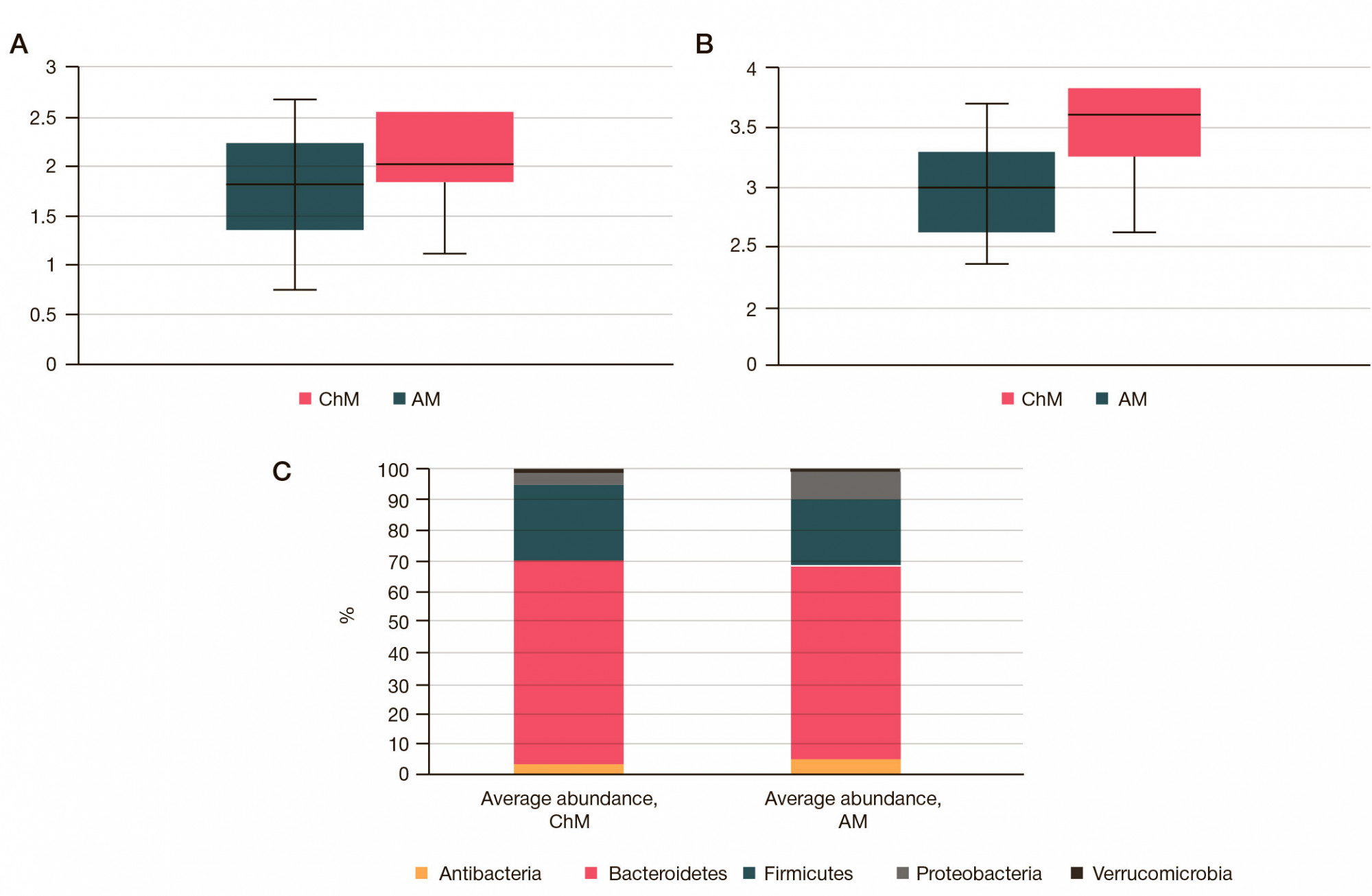
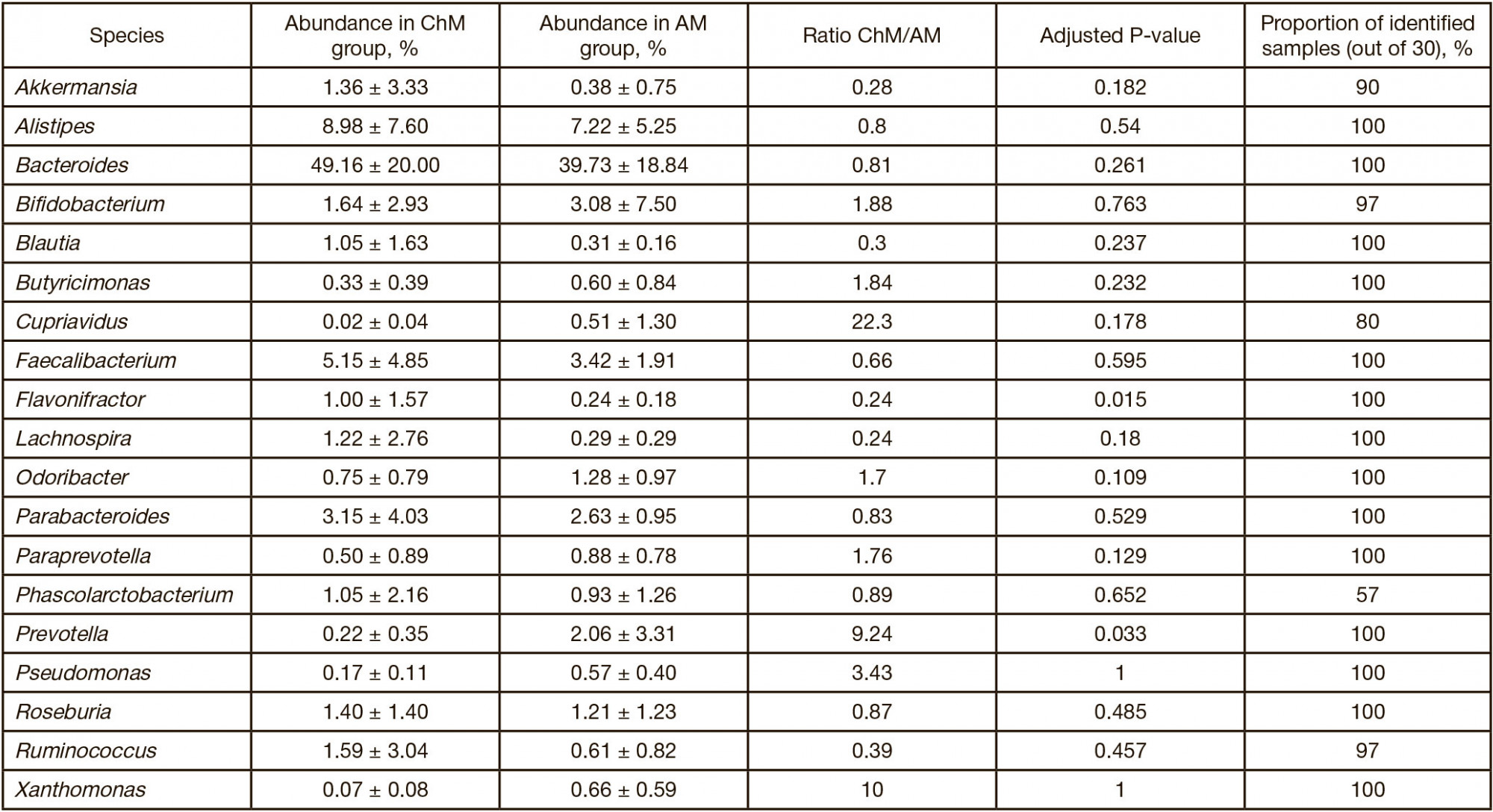
Altered neurometabolic potential of gut microbiome in healthy children of different age
1 Vavilov Institute of General Genetics, Moscow, Russia
2 Psychiatric Hospital № 1 Named after N. A. Alexeev, Moscow, Russia
Correspondence should be addressed: Alexey S. Kovtun
Gubkina, 3, Moscow, 119991; moc.liamg@52sanutvok
Funding: the study was supported by the Russian Science Foundation, project № 20-14-00132.
Compliance with ethical standards: the study was approved by the Ethics Committee of the Pirogov Russian National Research Medical University (protocol № 165 dated May 22, 2017). The informed consent was obtained from parents of all children.
Author contribution: Kovtun AS — algorithm development, bioinformatics analysis, catalogue creation, data interpretation and vizualization; Averina OV — method development, catalogue creation, data interpretation, manuscript writing; Poluektova EU — method development, catalogue creation; Kostyuk GP and Danilenko VN — study concept, method development, data interpretation.