This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (CC BY).
ORIGINAL RESEARCH
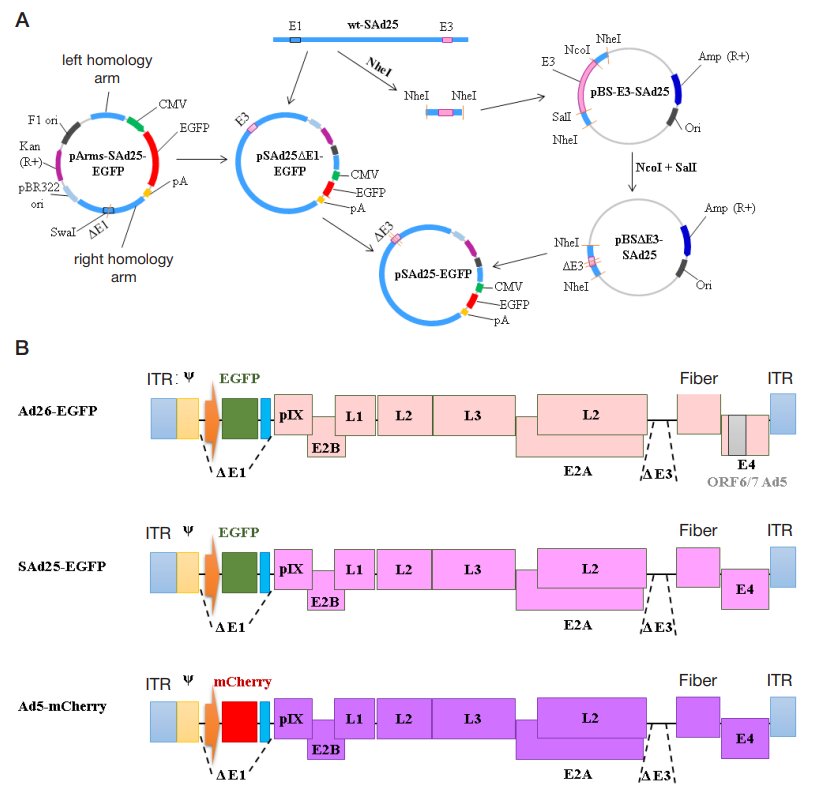
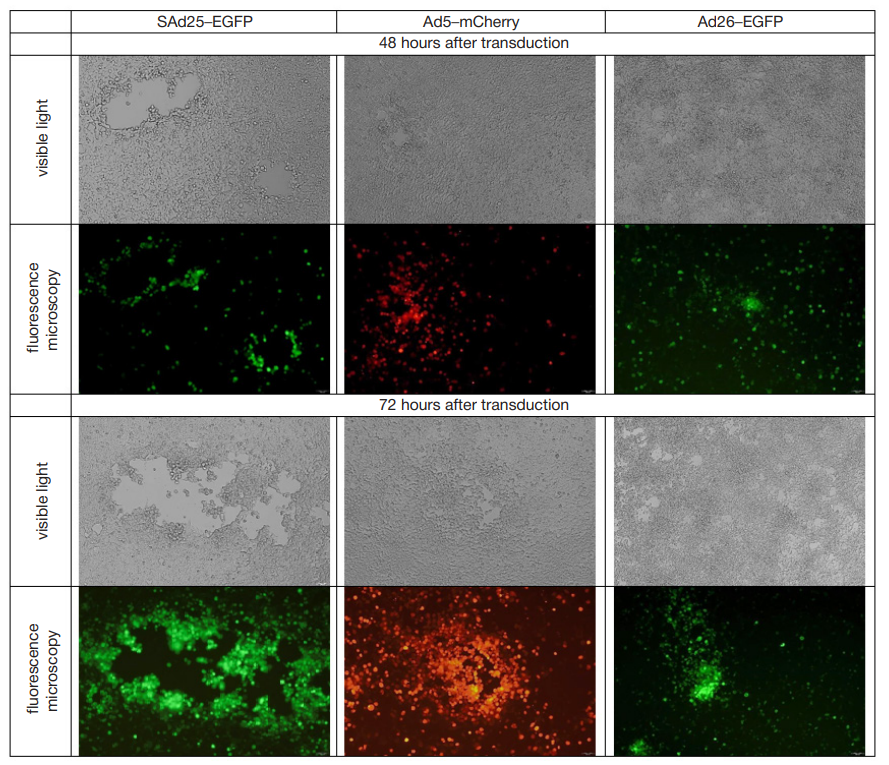
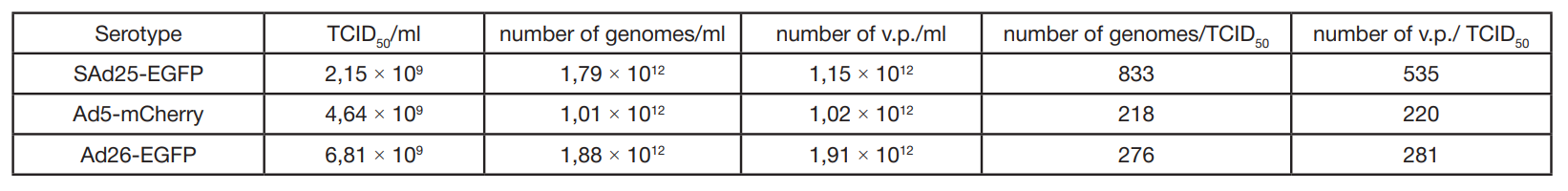
Development and characterization of a vector system based on the simian adenovirus type 25
Gamaleya National Research Center for Epidemiology and Microbiology of the Ministry of Health of the Russian Federation, Moscow, Russia
Correspondence should be addressed: Tatiana A. Ozharovskaia
Gamalei, 18, Moscow, 123098, Russia; ur.xednay@hzo.t
Funding: the work is the result of the effort under the "Development of a recombinant vaccines platform (including live vaccines) enabling design of the infectious diseases prevention vaccines" task set by the Ministry of Health of the Russian Federation.
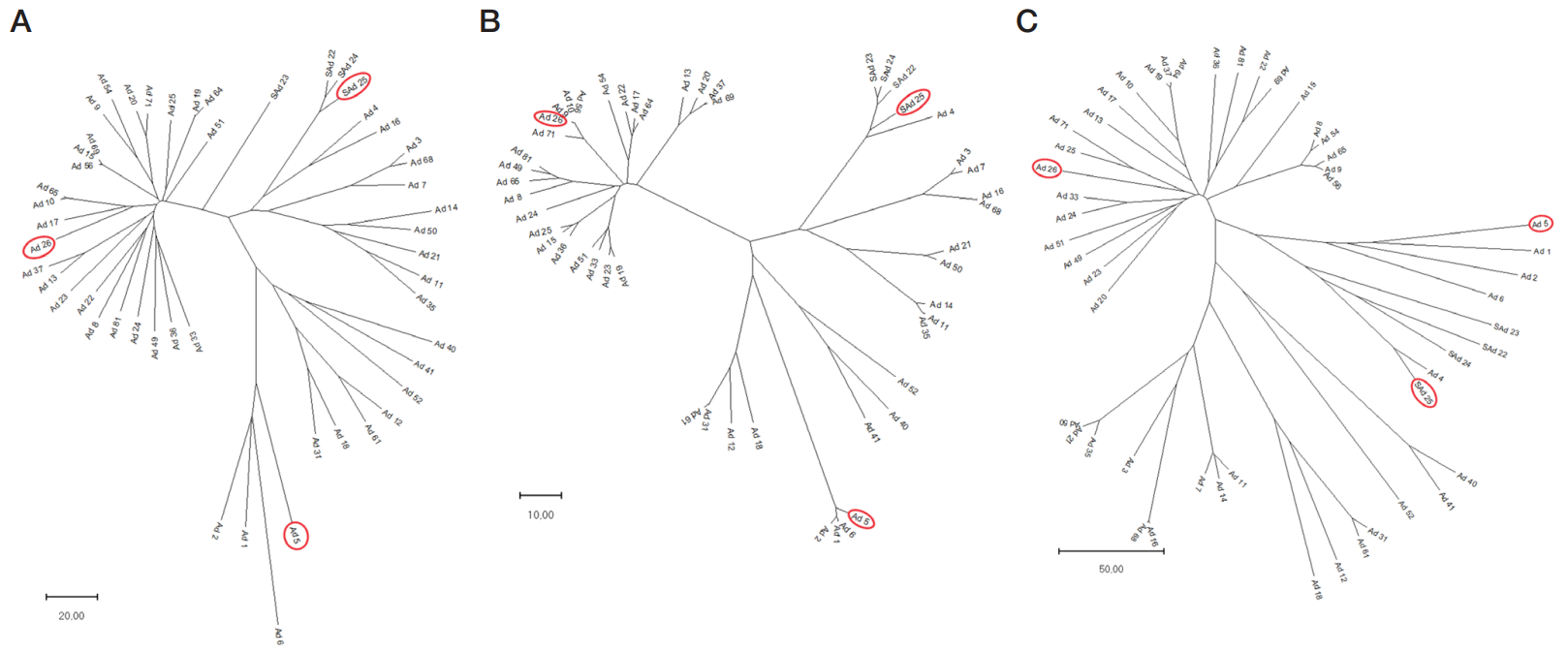
Acknowledgments: we express our sincere gratitude to AE Nikonova, a researcher with the Laboratory of Molecular Biotechnology of the NF Gamaleya National Research Center for Epidemiology and Microbiology for her help in conducting the phylogenetic analysis of adenovirus hexon sequences, as well as to research laboratory assistant D.D. Kustova for her help in obtaining the whole genome sequences.
Author contribution: Ozharovskaia TA, Zubkova OV, Logunov DYu, Gintsburg AL — conceptualization and planning of the experimental part; Popova O, Zubkova OV — production of genetically engineered constructs and recombinant simian adenovirus type 25; Ozharovskaia TA, Vavilova IV — phylogenetic analysis, virological experiments; Pochtovyi AA — whole genome sequencing of adenoviruses; Ozharovskaia TA, Zubkova OV — article preparation and authoring, interpretation of the results; Shcheblyakov DV, Gushchin VA — manuscript editing; Gushchin VA — state assignment project management.
Compliance with ethical standards: the work was carried out in accordance with the principles of the Declaration of Helsinki.