This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (CC BY).
ORIGINAL RESEARCH
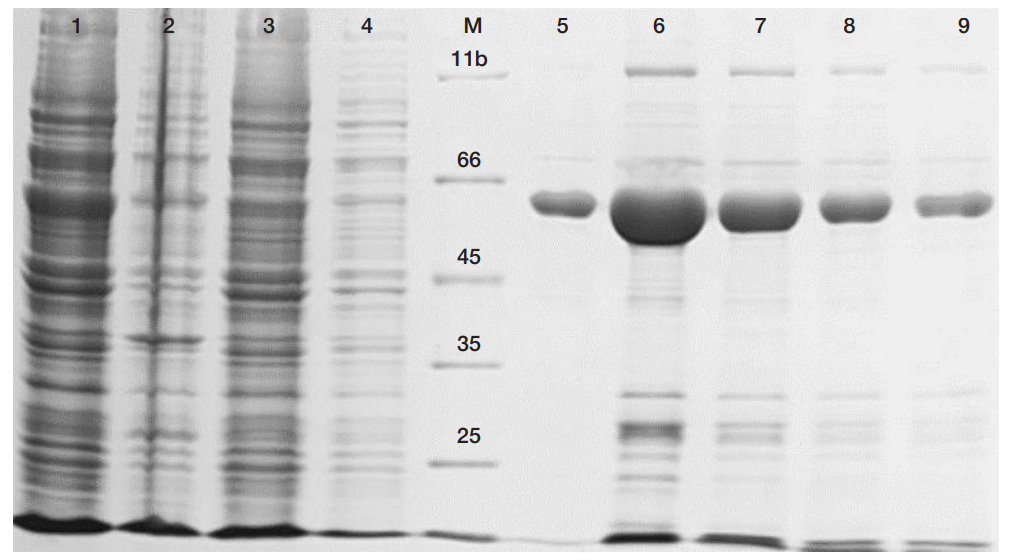
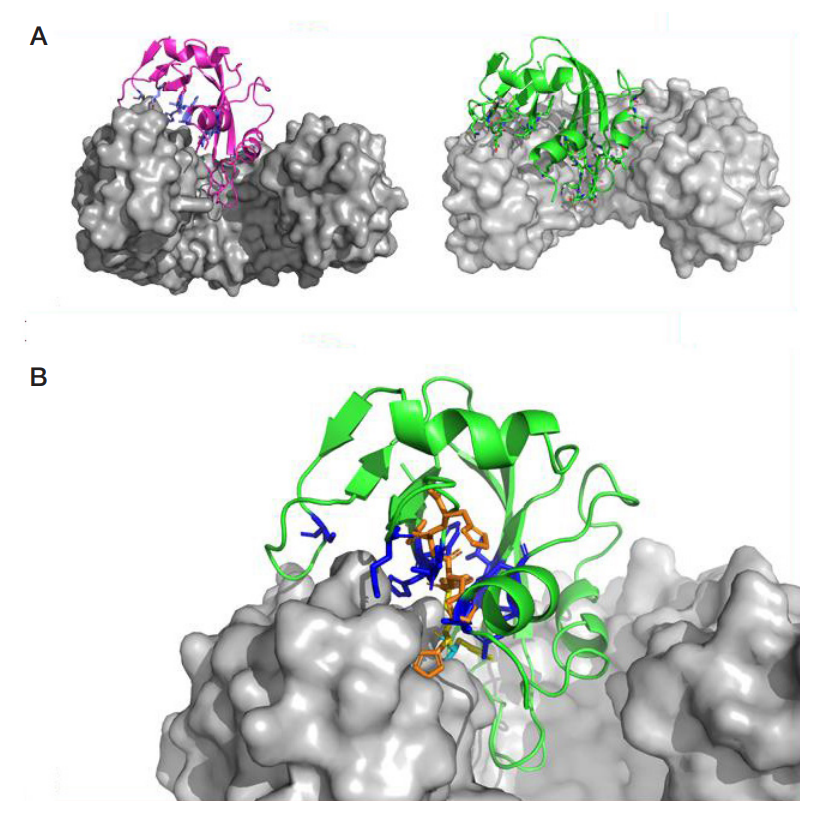
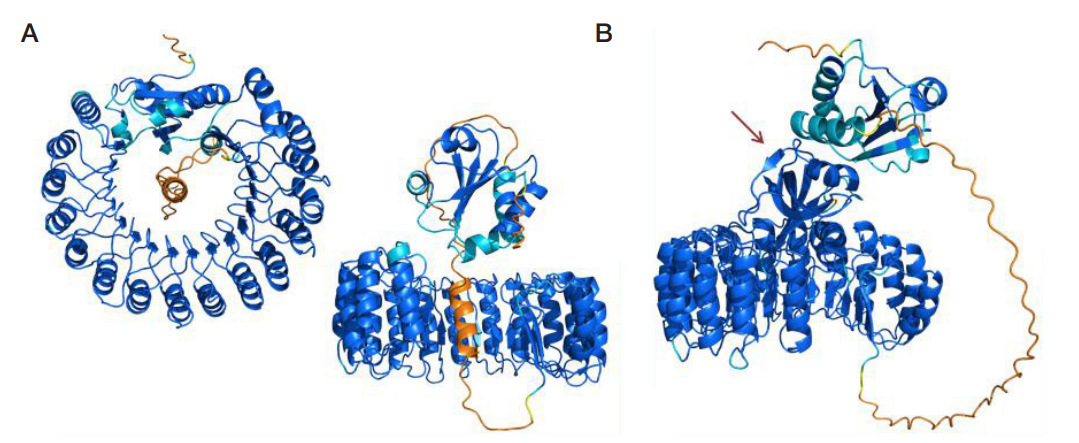
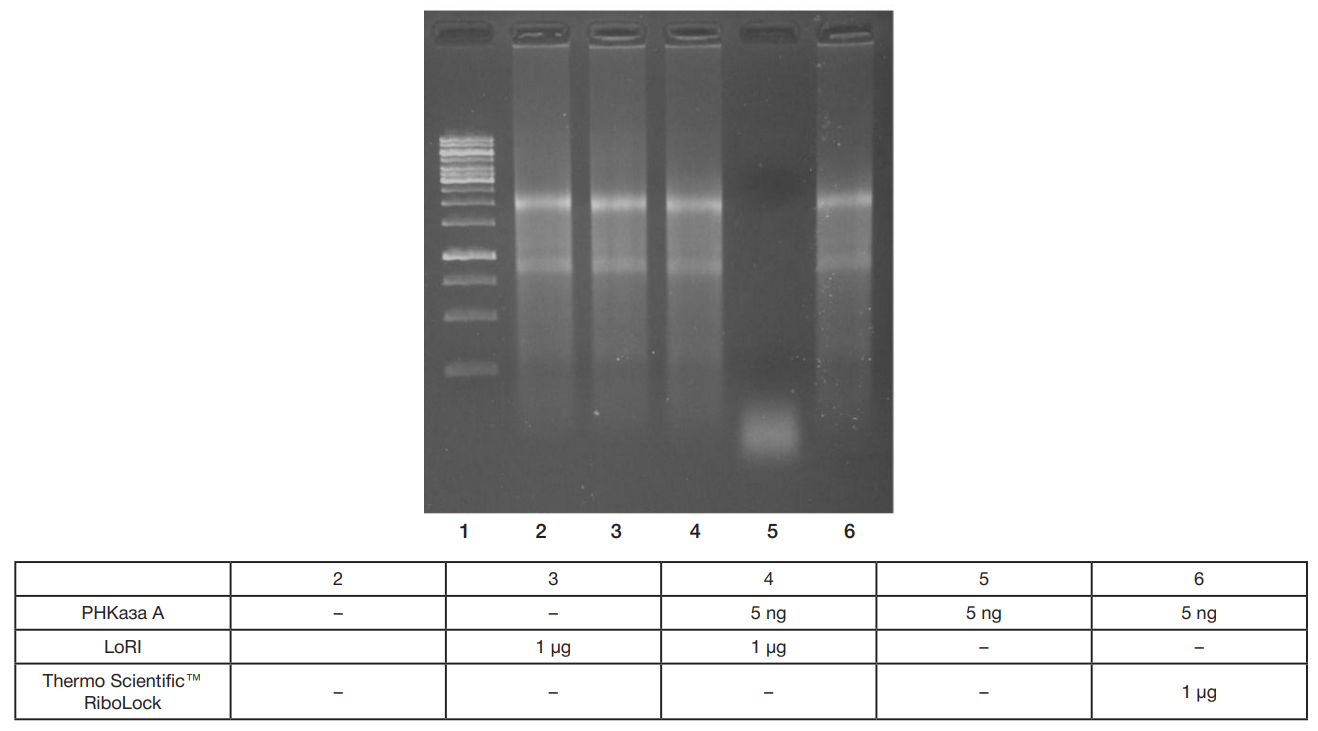
LoRI, a new recombinant RNase inhibitor for in vitro applications
1 DNA-Technology LLC, 117587, Moscow, Russia
2 MIREA — Russian Technological University, Moscow, Russia
3 Shemyakin–Ovchinnikov Institute of Bioorganic Chemistry RAS, Moscow, Russia
4 Bauman Moscow State Technical University, Moscow, Russia
5 Pirogov Russian National Research Medical University, Moscow, Russia
Correspondence should be addressed: Natalia Yu. Usman
Ostrovityanova 1/1, Moscow, 117997, Russia; ur.relbmar@namsu_n
Funding: The study was funded by DNA-Technology LLC.
Author contribution: Sukhov DA, Romanenko GA — protein engineering, expression and purification; Kholoshenko IV, Petrova TV — product characterization, writing; Myshkin MYu — protein structure analysis; Kost VYu — study design; Trofimov DYu — project supervision; Usman NYu — writing; Barsova EV — project coordination, writing.