This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (CC BY).
ORIGINAL RESEARCH
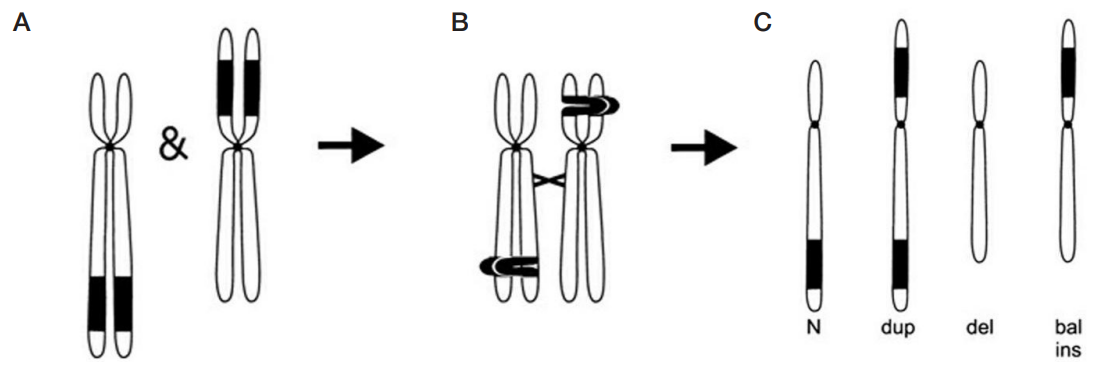
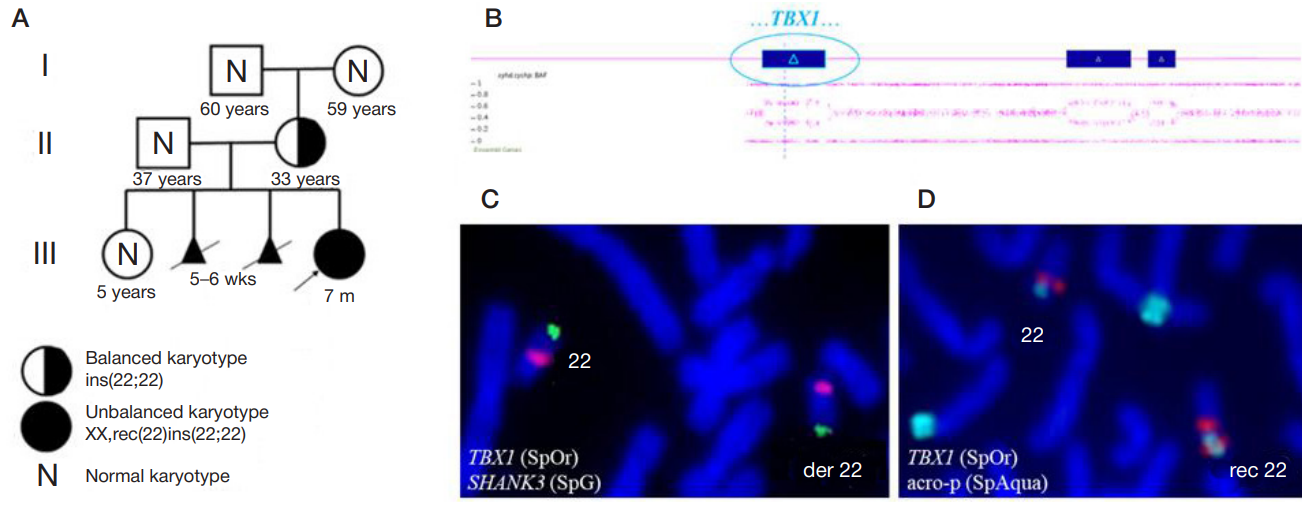
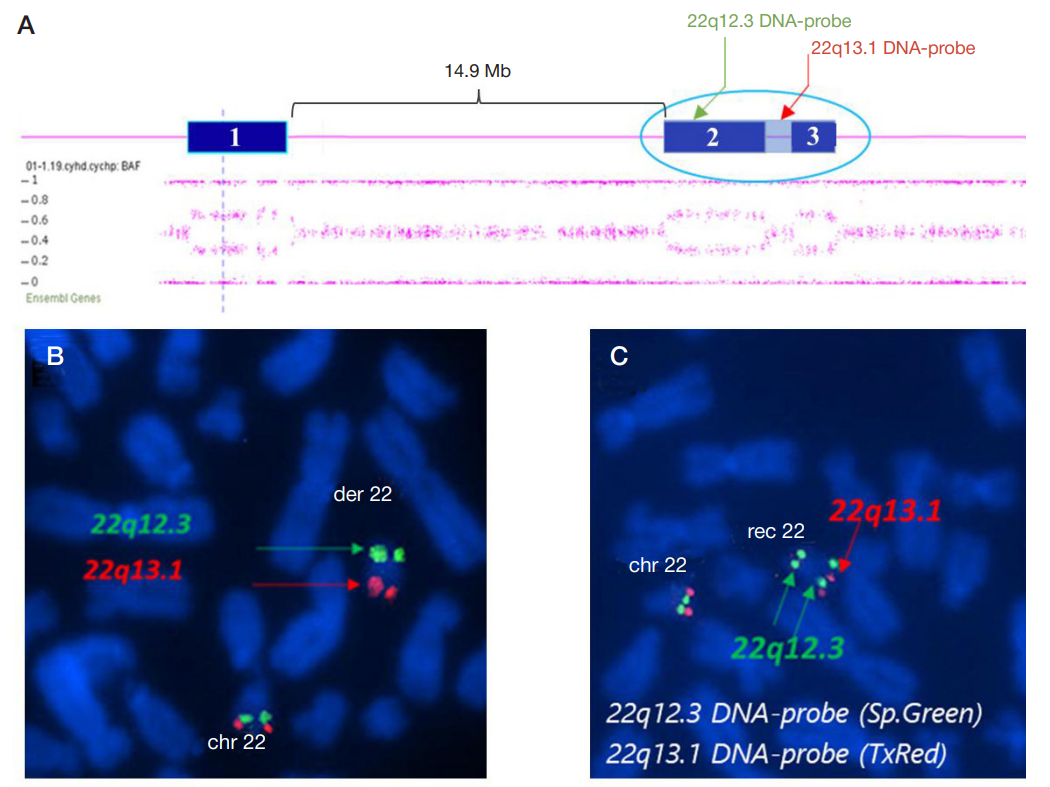
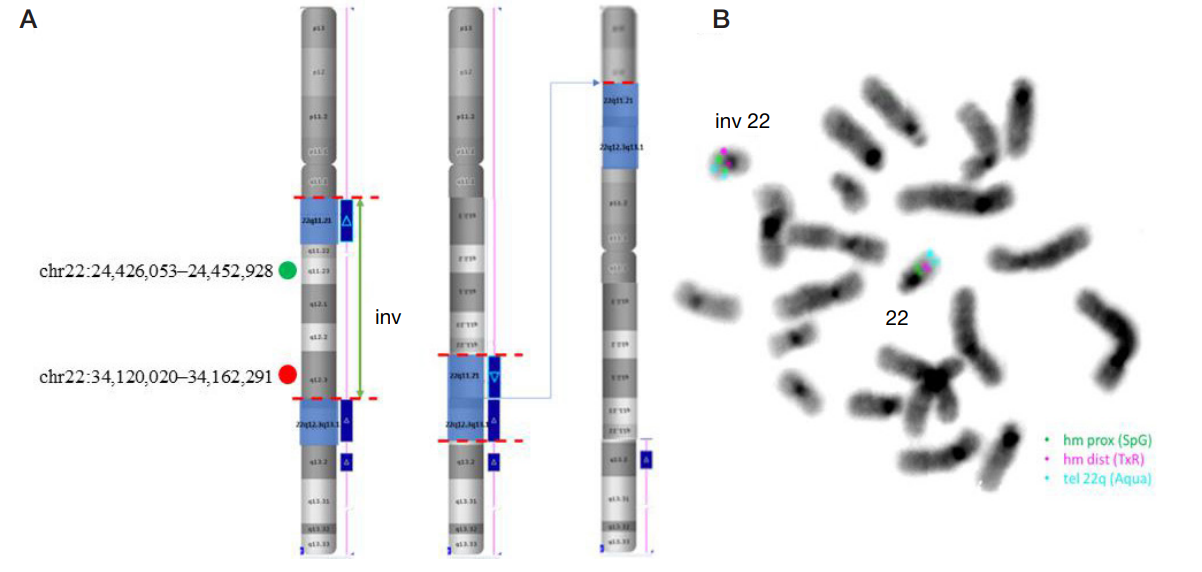
Molecular cytogenetic characterization of a rare recombinant chromosome 22 caused by a maternal intrachromosomal insertion
Research Centre for Medical Genetics, Moscow, Russia
Correspondence should be addressed: Darya A. Yurchenko
Moskvorechye, 1, Moscow, 115522, Russia; ur.liam@vblahsad
Funding: this research was supported by the Ministry of Science and Higher Education of the Russian Federation (#FGFF-2023-0003).
Author contribution: Yurchenko DA — study design, development of homemade DNA probes, conducting FISH analysis and interpreting the data, manuscript preparation; Markova ZhG — conducting FISH analysis using commercial DNA probes; Petukhova MS and Matyushchenko GN — clinical genetic counseling of the family; Shilova NV — study conception and design, discussion of results, and scientific editing of the manuscript.
Compliance with ethical standards: the study was approved by the Ethics Commitee of the Research Centre for Medical Genetics (protocol No. 4/2 dated 19 April 2021). The informed consent for participation in the research study was obtained from the patients.