This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (CC BY).
ORIGINAL RESEARCH
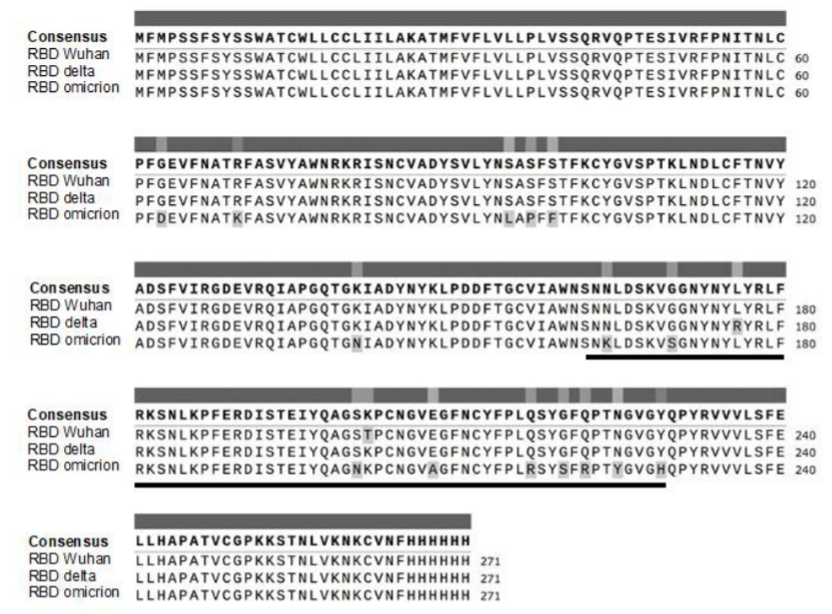
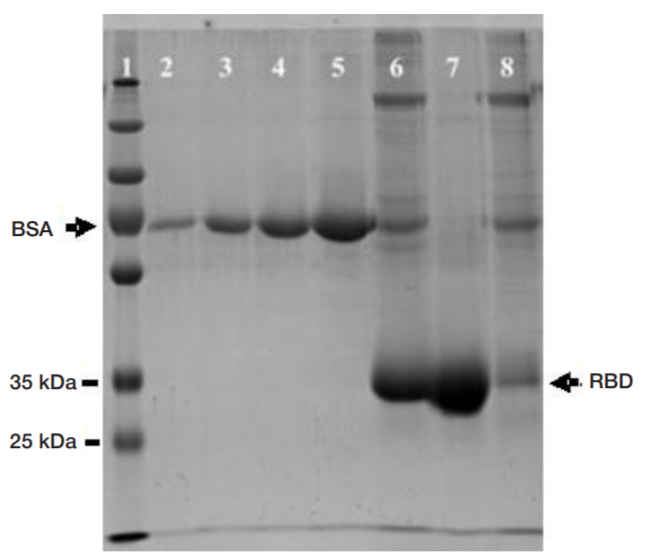
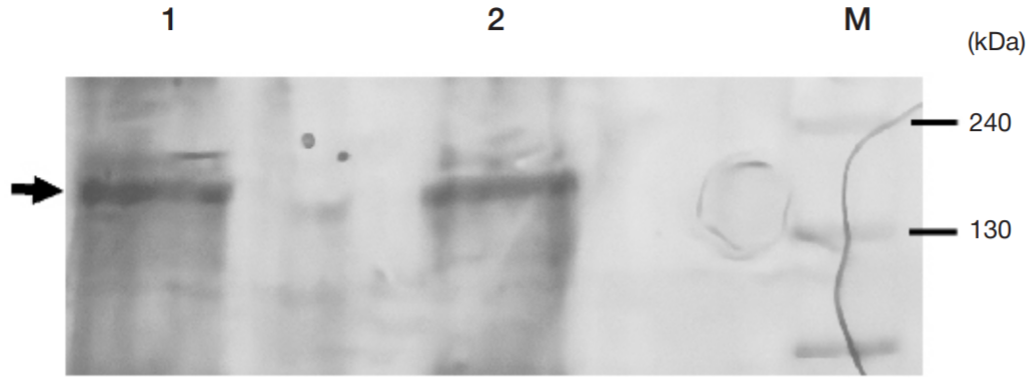
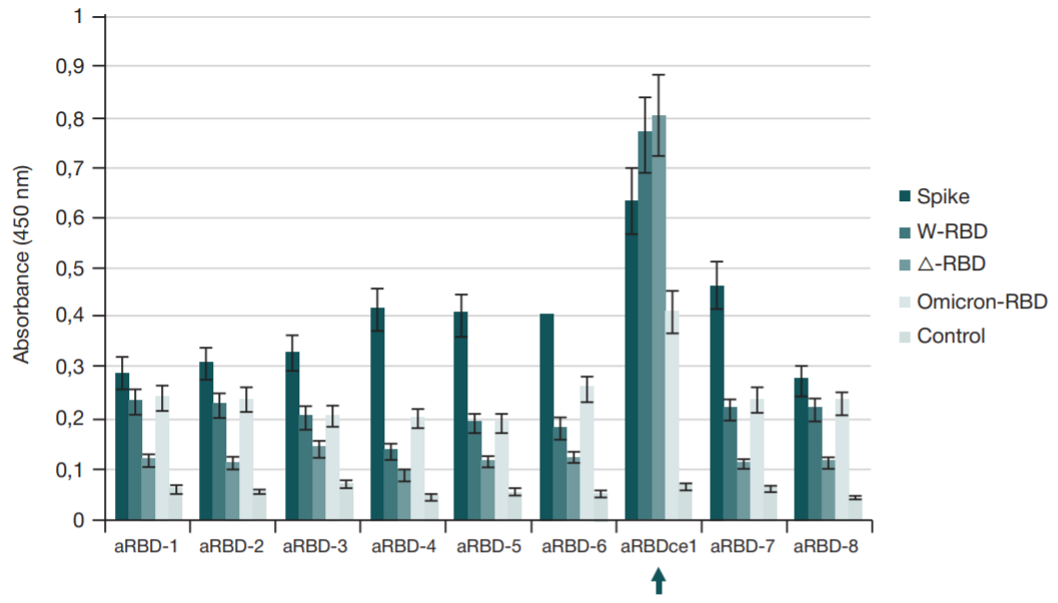
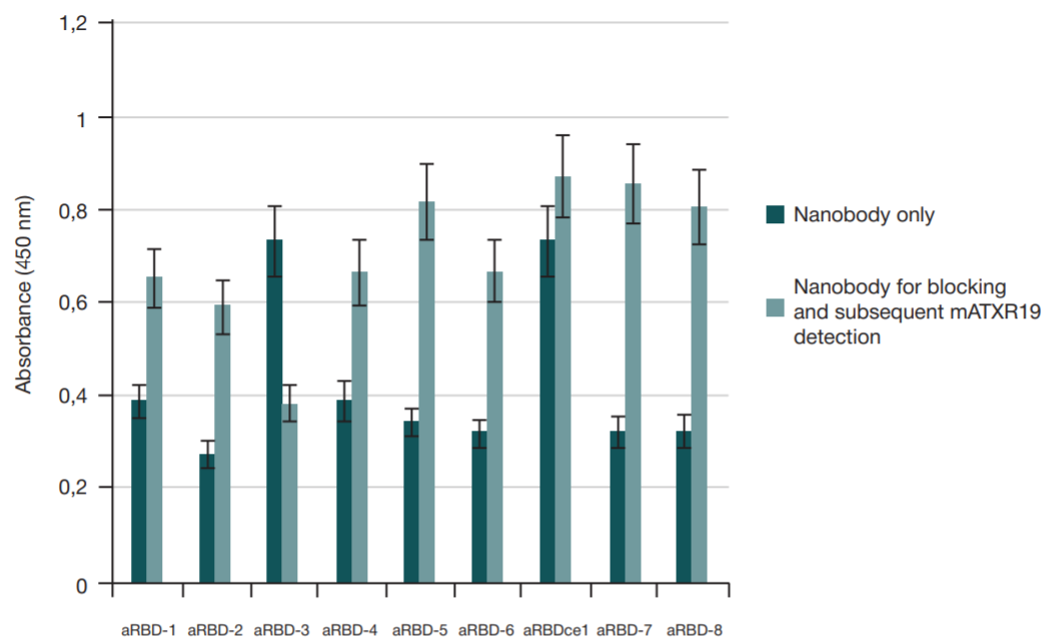
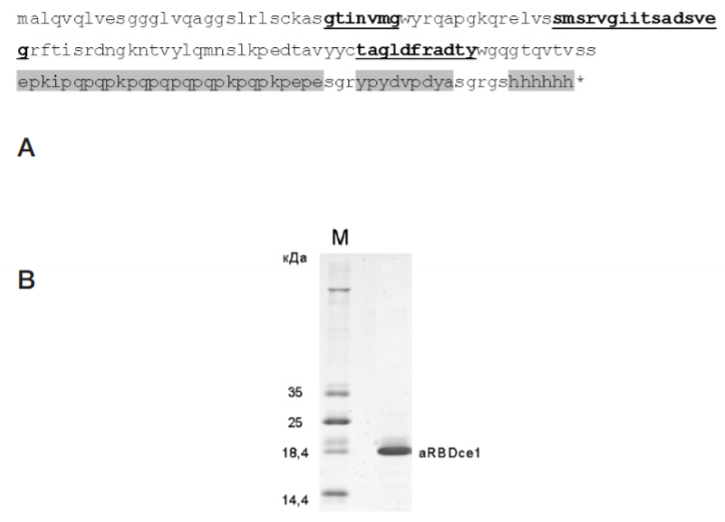
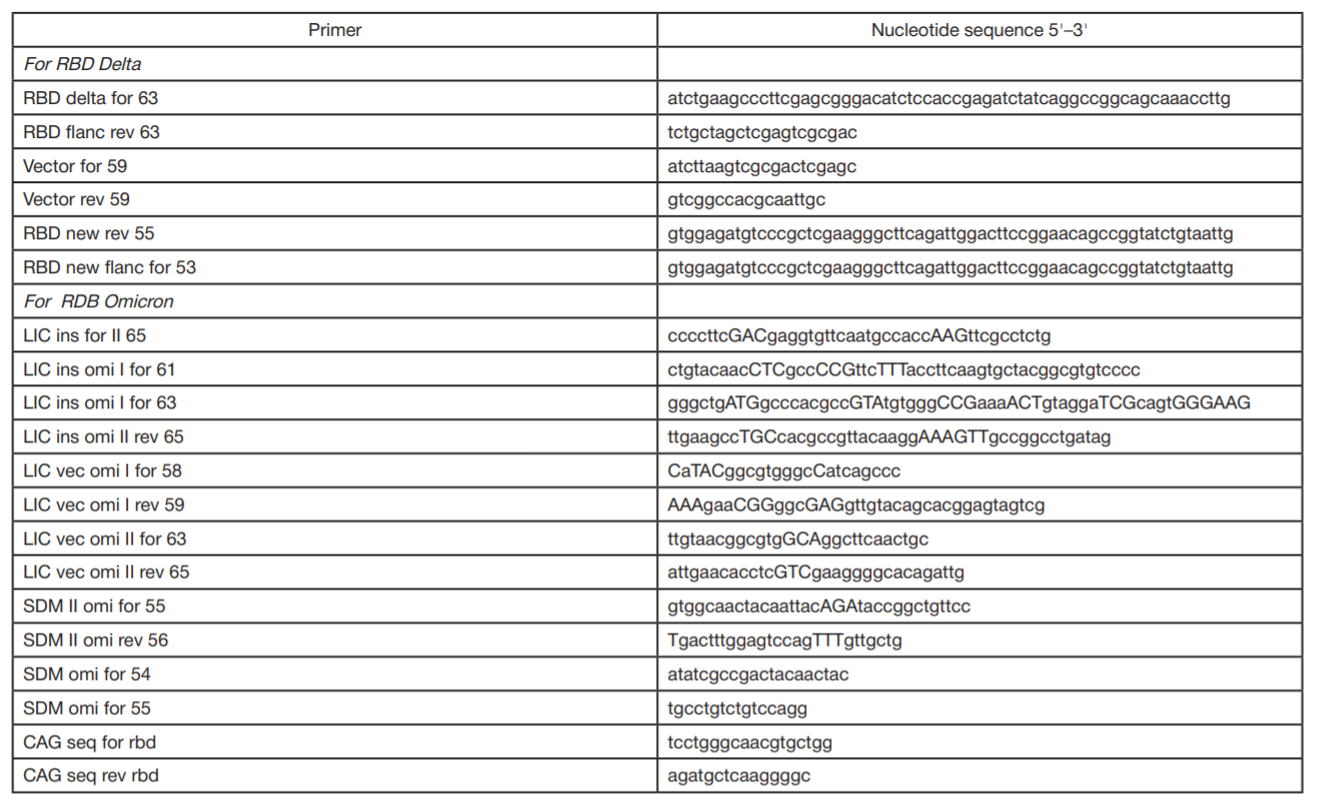
Single-domain antibody for binding the conserved epitope in the SARS-CoV-2 spike protein receptor-binding domain
1 Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, Moscow, Russia
2 Institute of Gene Biology, Russian Academy of Sciences, Moscow, Russia
Correspondence should be addressed: Sergei V. Tillib
Vavilov str., 34/5, Moscow, 119334, Russia; ur.ygoloibeneg@billit moc.liamg@billit.iegres
Funding: the study was supported by the Ministry of Science and Higher Education of the Russian Federation (agreement № 075-15-2021-1086, contract № RF––193021X0015).
Acknowledgements: the authors thank M.V. Rutovskaya, Severtsov Institute of Ecology and Evolution of the Russian Academy of Sciences, for assistance in immunization of the camel.
Author contribution: Vorobyev PO — molecular cloning and subsequent production of recombinant proteins (antigens for immunization); Tillib SV — developing general conception, carrying out immunization, developing the method for acquisition and primary analysis of the generated single-domain antibodies, manuscript writing.
Compliance with ethical standards: the study was approved by the Ethics Committee of the Severtsov Institute of Ecology and Evolution of the Russian Academy of Sciences (protocol № 17 of 11 February 2018); the animal was handled in strict compiance with the guidelines of the National Standard of the Russian Federation GOST R 53434–2009.