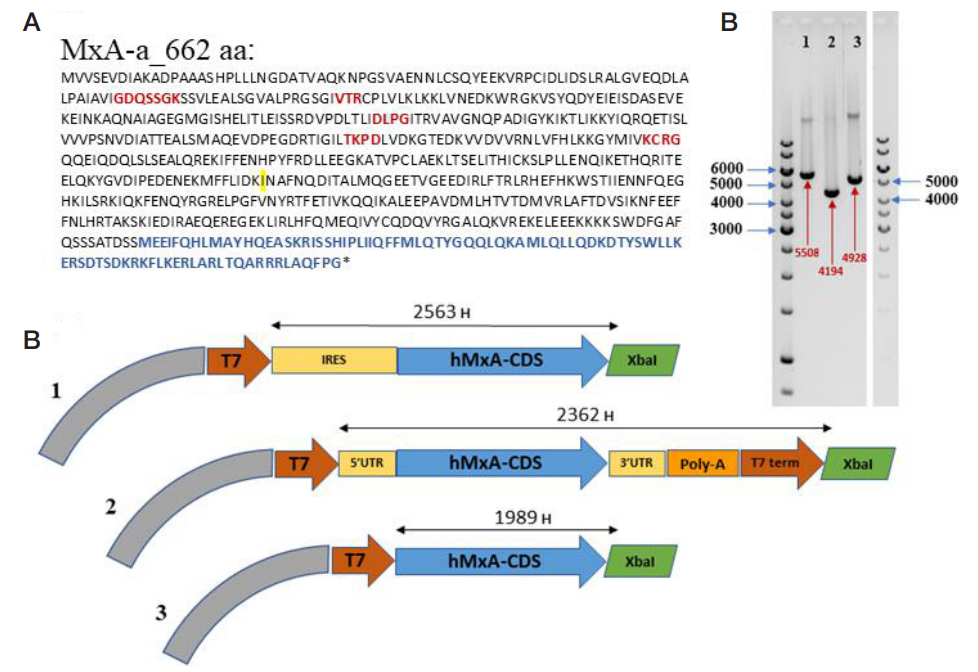
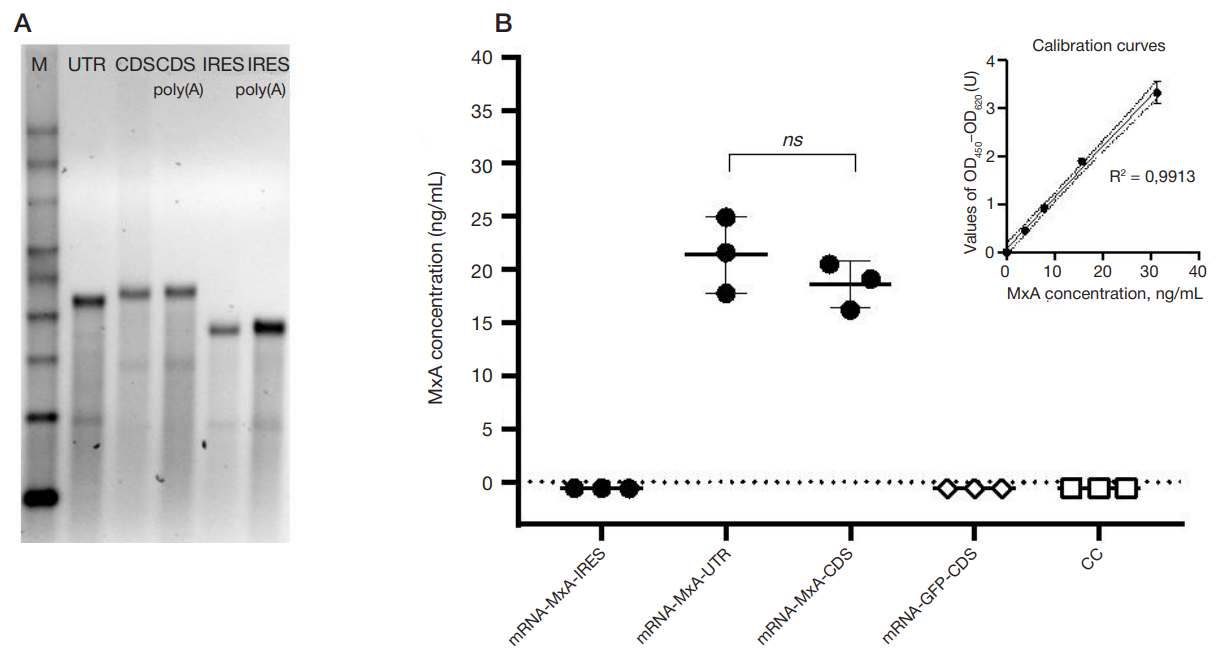
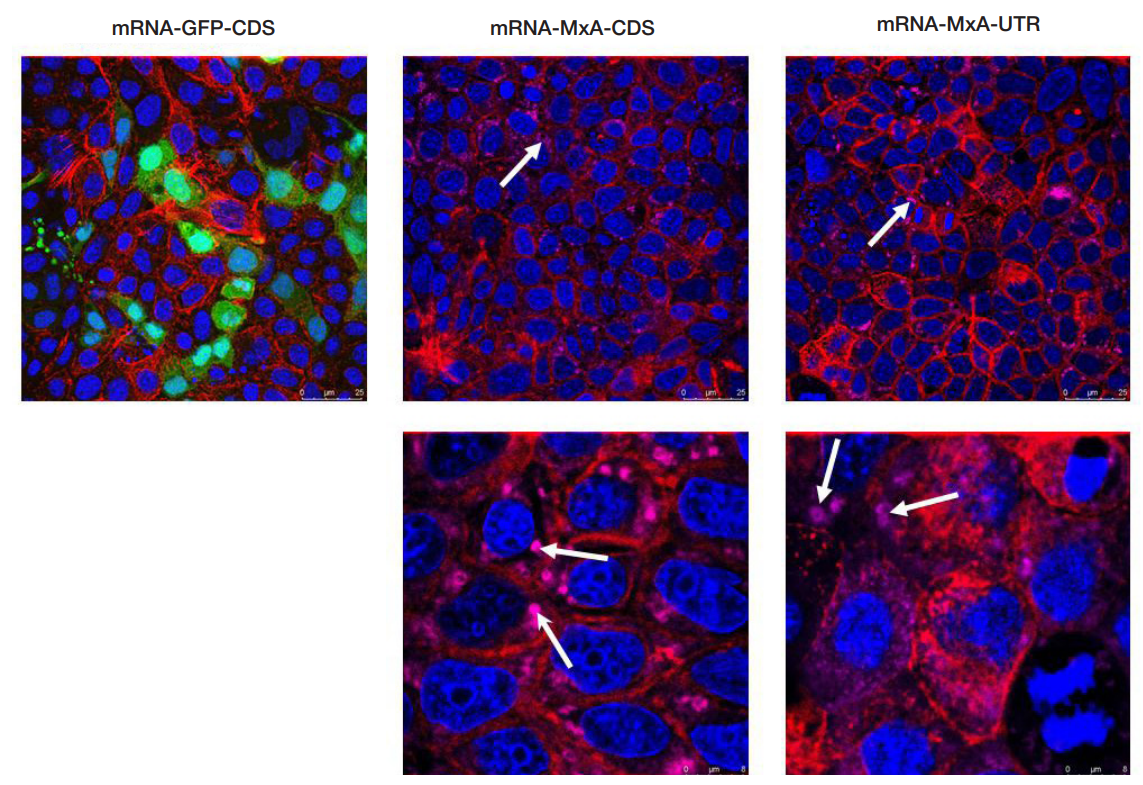
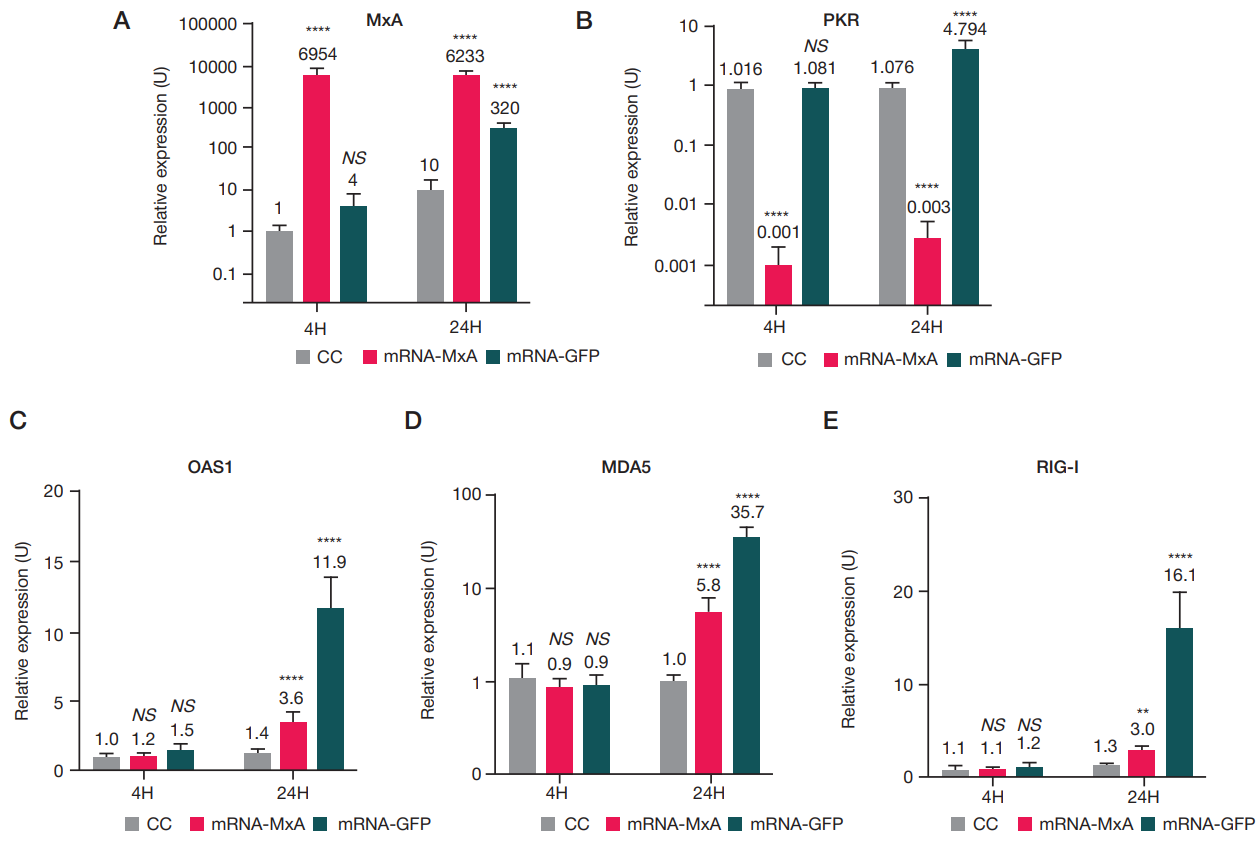
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (CC BY).
ORIGINAL RESEARCH
Production and biological activity of the exogenous mRNA encoding human MxA protein
1 Smorodintsev Research Institute of Influenza of the Ministry of Health of the Russian Federation, St. Petersburg, Russia
2 Institute of Cytology, Russian Academy of Science, St. Petersburg, Russia
Correspondence should be addressed: Sergey A. Klotchenko
Professora Popova, 15/17, St. Petersburg, 197022, Russia; ur.liam@kitafsof
Funding: the study was supported by the Russian Science Foundation, Agreement No. 23-25-00433, project title: “Assessment of antiviral effect of the mRNA encoding human MxA protein” (manager M.A. Plotnikova), https://rscf.ru/project/23-25-00433/
Author contribution: Plotnikova MA — study design, experimental procedure, analysis of the results, statistical processing, manuscript writing; Bobkov DE — confocal microscopy examination; Klotchenko SA — production and characterization of the exogenous mRNA preparations, manuscript editing.