This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (CC BY).
ORIGINAL RESEARCH
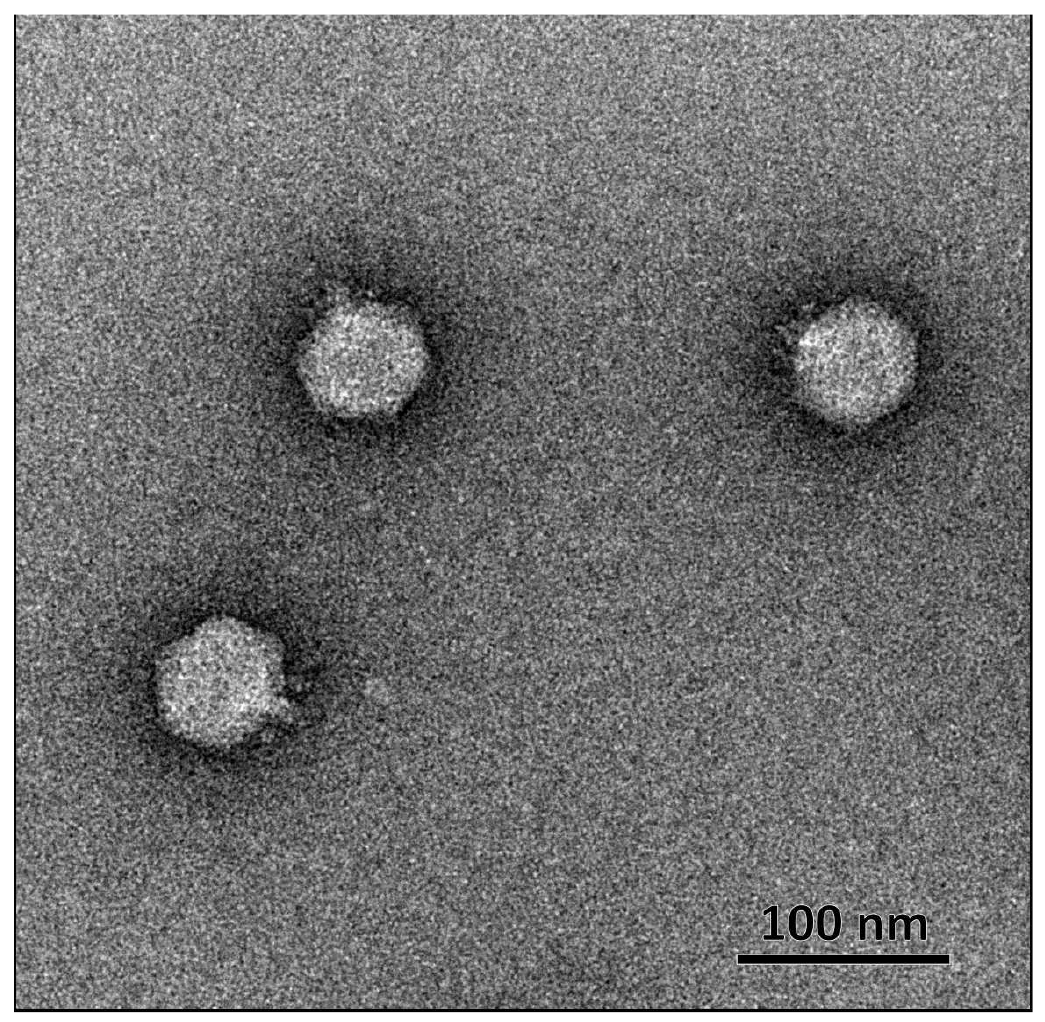
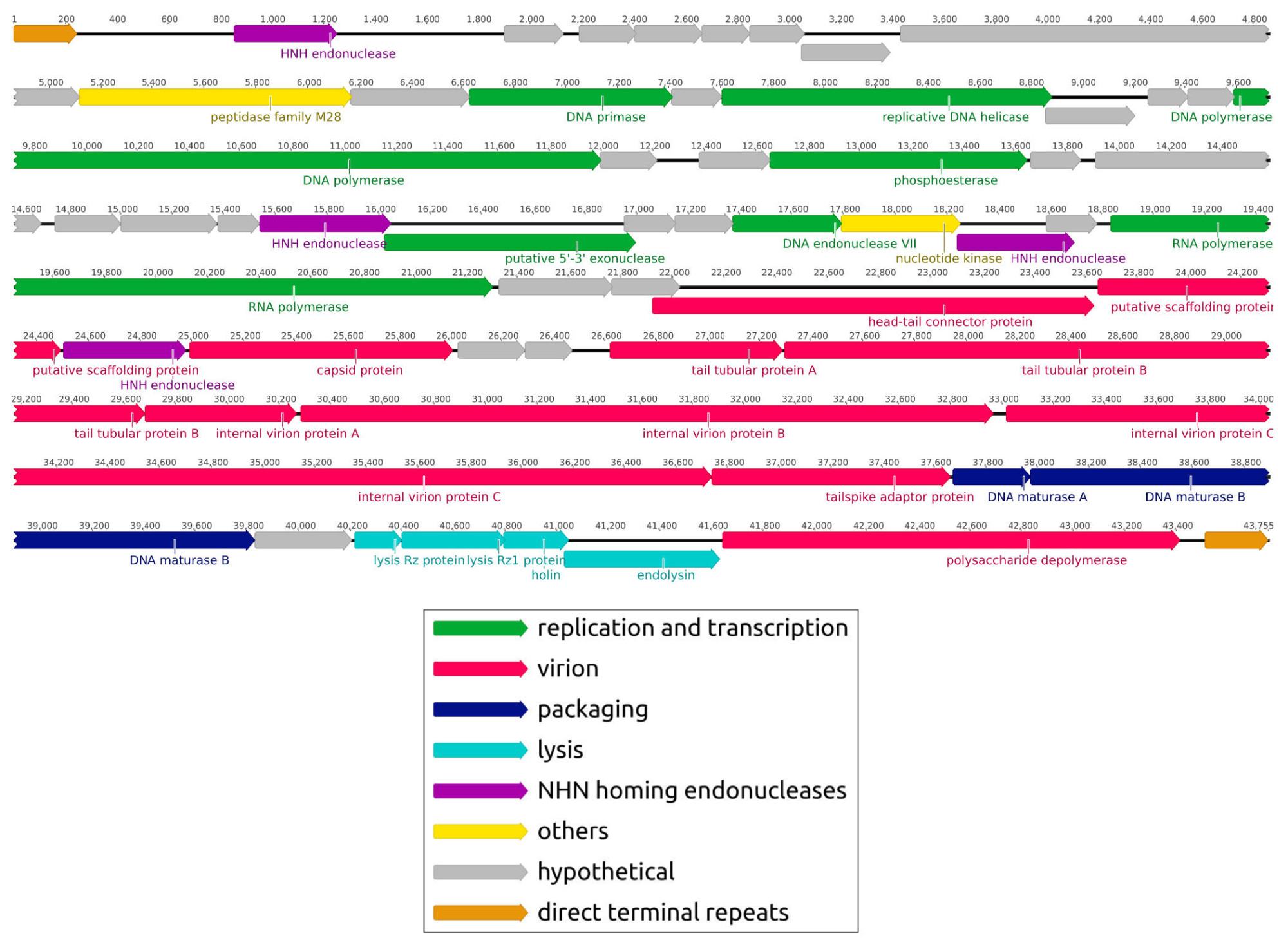
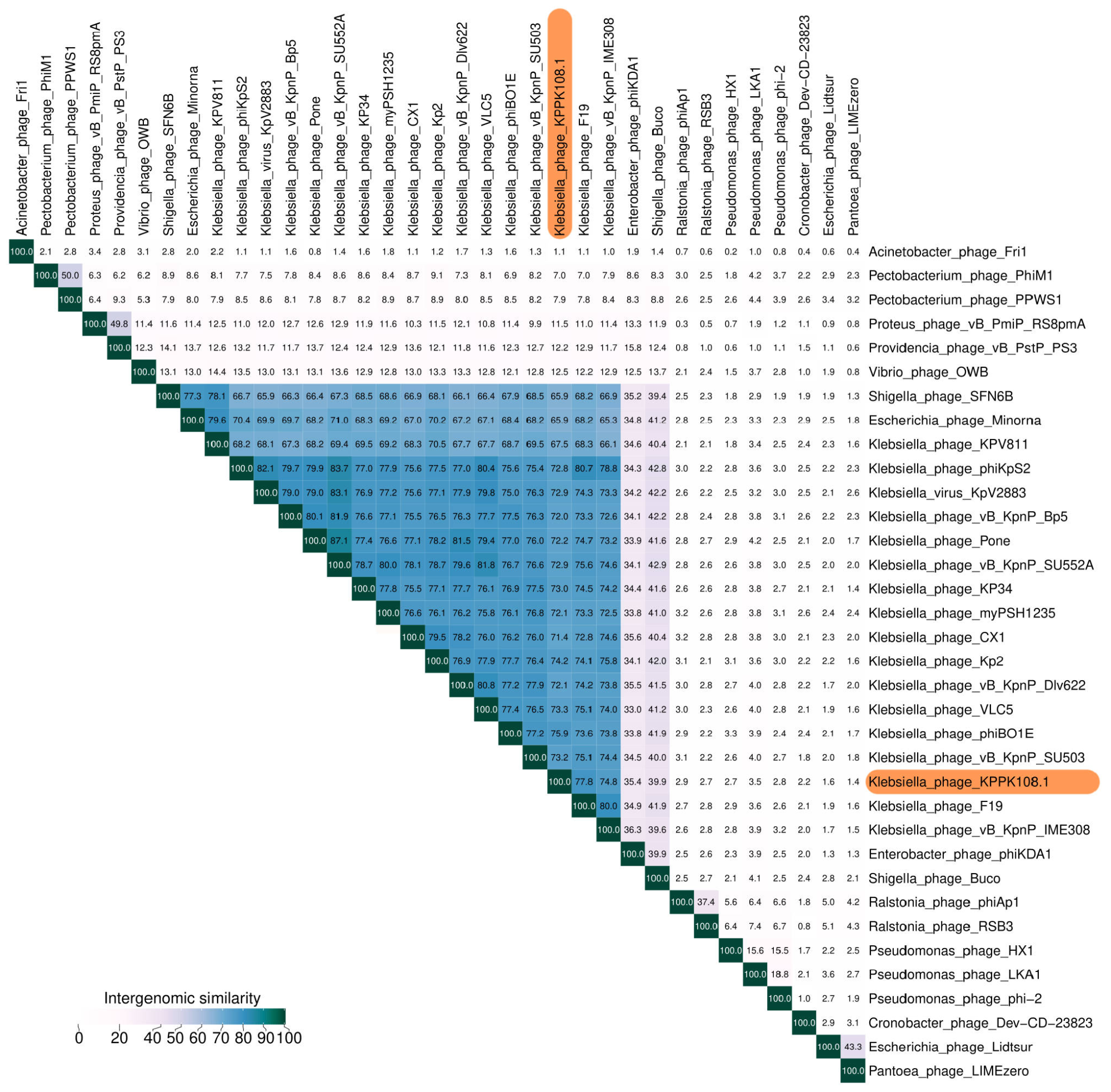
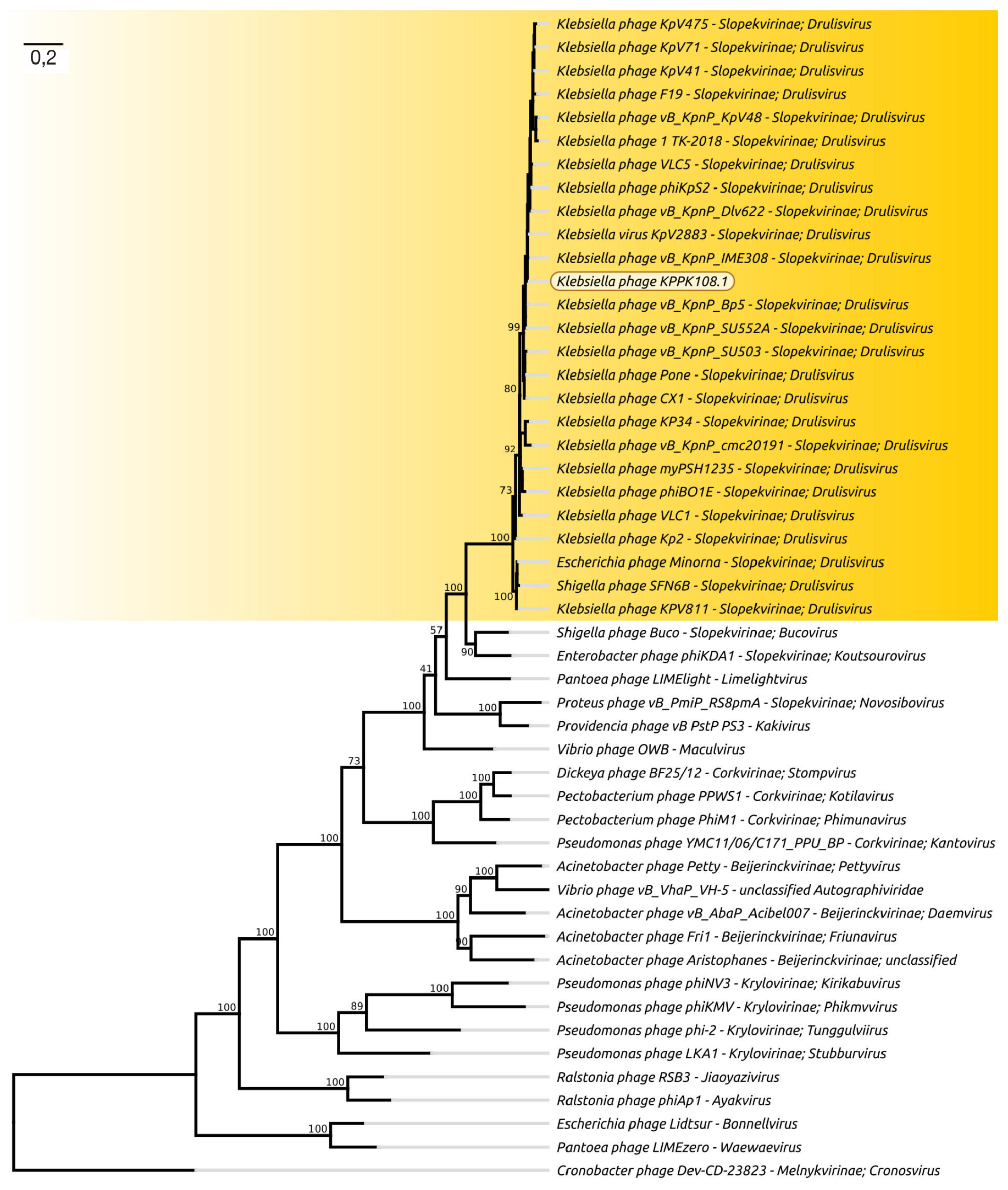
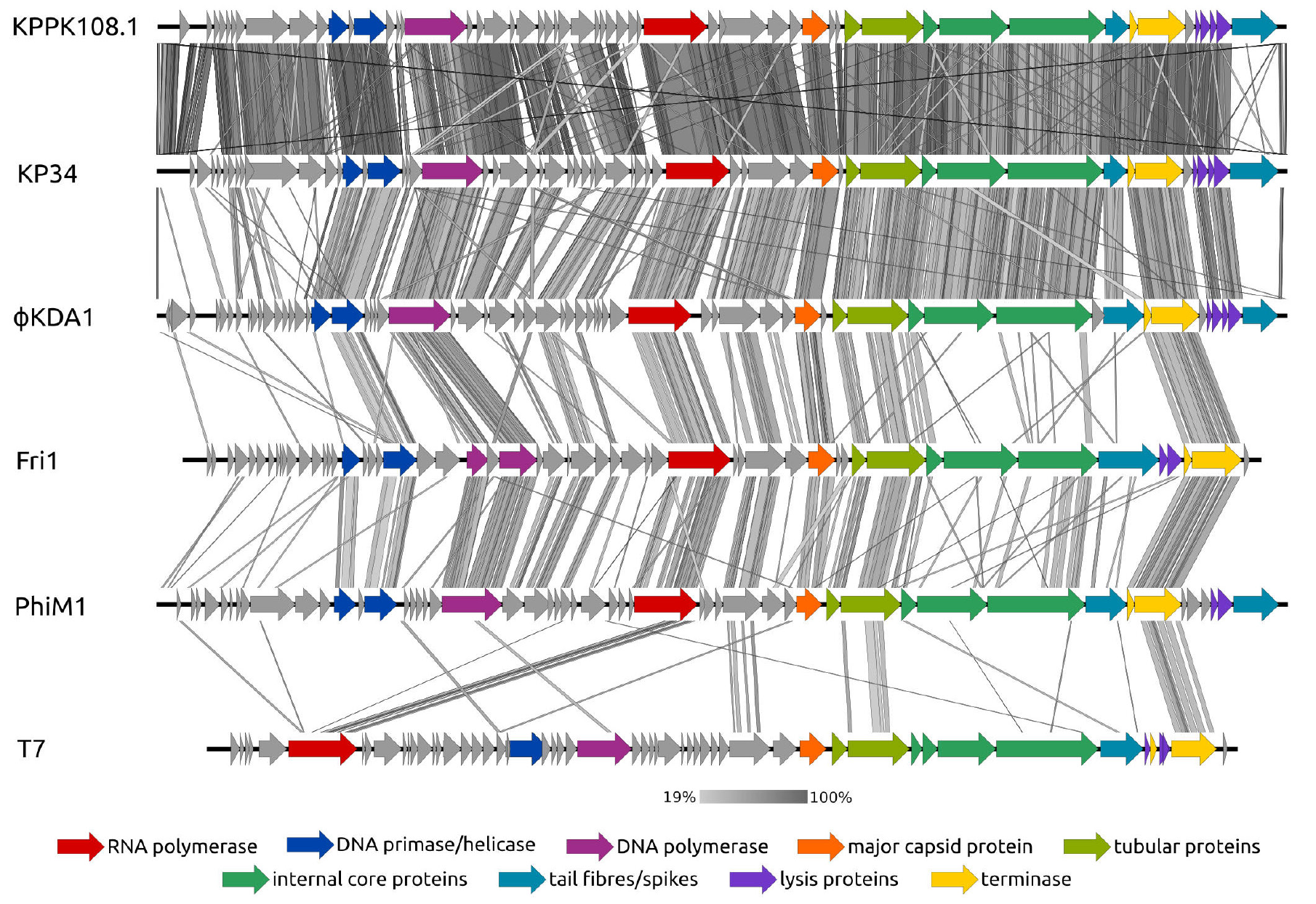
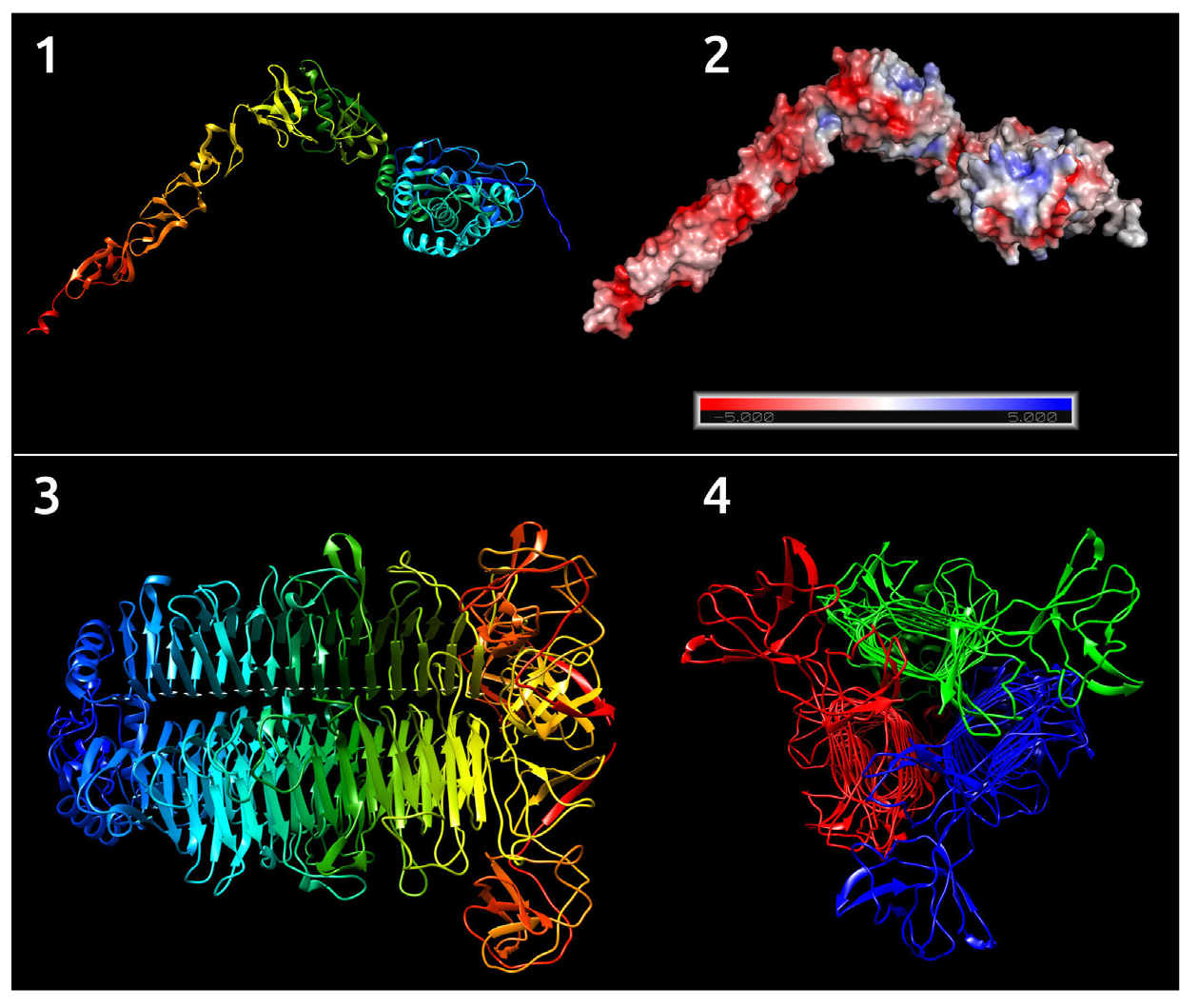
Novel Klebsiella pneumoniae virulent bacteriophage KPPK108.1 capable of infecting the K108 serotype strains
1 Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Moscow, Russia
2 Central Research Institute of Epidemiology of Rospotrebnadzor, Moscow, Russia
3 Pirogov Russian National Research Medical University, Moscow, Russia
4 Lomonosov Moscow State University, Moscow, Russia
Correspondence should be addressed: Mikhail M. Shneider
Miklukho-Maklaya, 16/10, Moscow, 117997, Russia; ur.liam@nhs_mm
Funding: the study was funded by the Ministry of Health of the Russian Federation (EGISU R&D № 121052800048-3).
Acknowledgements: the authors wish to thank the Center for Precision Genome Editing and Genetic Technologies for Biomedicine, Pirogov Russian National Research Medical University, for advice on research methods.
Author contribution: Shagin DA — research conceptualization, study management, manuscript writing; Evseev PV, Shelenkov AA — formal analysis of sequencing data, manuscript editing; Shneider MM — methodology, study management; Mikhailova YuV — sequencing, data validation; Yanushevich YuG, Moiseenko AV, Karlova MG — methodology; Sokolova OS — electron microscopy, methodology.